Q9ZQR3
Gene name |
At2g14510 (T13P21.11) |
Protein name |
Leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14510 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT2G14510 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
704-711 (Activation loop from InterPro)
Target domain |
563-835 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
722-729 (Activation loop from InterPro)
Target domain |
563-835 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
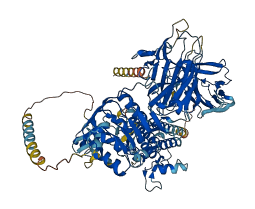
Autoinhibited structure

Activated structure

1 structures for Q9ZQR3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9ZQR3-F1 | Predicted | AlphaFoldDB |
215 variants for Q9ZQR3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH13090661 | 2 | E>K | No | 1000Genomes | |
| ENSVATH05460840 | 5 | N>S | No | 1000Genomes | |
| ENSVATH01786960 | 7 | F>L | No | 1000Genomes | |
| tmp_2_6175030_A_G | 8 | M>T | No | 1000Genomes | |
| ENSVATH05460839 | 16 | S>A | No | 1000Genomes | |
| ENSVATH05460838 | 18 | M>L | No | 1000Genomes | |
| tmp_2_6174997_G_T | 19 | S>Y | No | 1000Genomes | |
| ENSVATH01786958 | 26 | Q>L | No | 1000Genomes | |
| ENSVATH01786957 | 27 | Q>E | No | 1000Genomes | |
| ENSVATH01786945 | 39 | K>N | No | 1000Genomes | |
| ENSVATH13090624 | 40 | E>K | No | 1000Genomes | |
| tmp_2_6174821_G_A | 41 | S>F | No | 1000Genomes | |
| ENSVATH01786943 | 43 | I>T | No | 1000Genomes | |
| ENSVATH01786942 | 46 | S>A | No | 1000Genomes | |
| tmp_2_6174806_G_A | 46 | S>L | No | 1000Genomes | |
| ENSVATH01786941 | 47 | S>N | No | 1000Genomes | |
| ENSVATH01786941 | 47 | S>T | No | 1000Genomes | |
| tmp_2_6174801_T_A | 48 | N>Y | No | 1000Genomes | |
| ENSVATH01786939 | 49 | L>I | No | 1000Genomes | |
| ENSVATH07863183 | 51 | F>Y | No | 1000Genomes | |
| tmp_2_6174776_T_C | 56 | N>S | No | 1000Genomes | |
| ENSVATH05460828 | 57 | F>L | No | 1000Genomes | |
| ENSVATH01786936 | 67 | Q>E | No | 1000Genomes | |
| ENSVATH01786935 | 75 | I>L | No | 1000Genomes | |
| tmp_2_6174719_A_C | 75 | I>S | No | 1000Genomes | |
| tmp_2_6174698_A_C | 82 | L>W | No | 1000Genomes | |
| ENSVATH00228631 | 89 | I>S | No | 1000Genomes | |
| ENSVATH13090623 | 90 | R>* | No | 1000Genomes | |
| ENSVATH13090622 | 90 | R>K | No | 1000Genomes | |
| ENSVATH13090621 | 92 | C>R | No | 1000Genomes | |
| ENSVATH07863180 | 96 | S>N | No | 1000Genomes | |
| tmp_2_6174655_A_T | 96 | S>R | No | 1000Genomes | |
| ENSVATH01786930 | 98 | K>E | No | 1000Genomes | |
| ENSVATH01786929 | 100 | G>S | No | 1000Genomes | |
| ENSVATH01786928 | 103 | Y>F | No | 1000Genomes | |
| ENSVATH05460827 | 105 | I>M | No | 1000Genomes | |
| ENSVATH05460826 | 110 | Y>F | No | 1000Genomes | |
| ENSVATH14447636 | 117 | L>V | No | 1000Genomes | |
| ENSVATH13090620 | 118 | N>D | No | 1000Genomes | |
| ENSVATH00228630 | 119 | T>I | No | 1000Genomes | |
| tmp_2_6174565_A_C | 126 | F>L | No | 1000Genomes | |
| ENSVATH01786923 | 133 | T>K | No | 1000Genomes | |
| ENSVATH05460825 | 134 | S>I | No | 1000Genomes | |
| ENSVATH14447635 | 136 | D>N | No | 1000Genomes | |
| ENSVATH07863177 | 139 | I>K | No | 1000Genomes | |
| ENSVATH01786922 | 141 | D>G | No | 1000Genomes | |
| ENSVATH07863176 | 142 | V>G | No | 1000Genomes | |
| ENSVATH01786920 | 151 | V>I | No | 1000Genomes | |
| ENSVATH13090617 | 169 | S>T | No | 1000Genomes | |
| ENSVATH01786915 | 176 | I>V | No | 1000Genomes | |
| ENSVATH05460823 | 188 | A>T | No | 1000Genomes | |
| ENSVATH07863175 | 189 | R>Q | No | 1000Genomes | |
| tmp_2_6174362_T_C | 194 | K>R | No | 1000Genomes | |
| ENSVATH09456760 | 195 | S>N | No | 1000Genomes | |
| ENSVATH09456760 | 195 | S>T | No | 1000Genomes | |
| ENSVATH09456759 | 196 | M>I | No | 1000Genomes | |
| tmp_2_6174357_T_G | 196 | M>L | No | 1000Genomes | |
| ENSVATH13090616 | 197 | A>E | No | 1000Genomes | |
| ENSVATH13090616 | 197 | A>V | No | 1000Genomes | |
| ENSVATH01786911 | 198 | H>L | No | 1000Genomes | |
| ENSVATH01786911 | 198 | H>R | No | 1000Genomes | |
| tmp_2_6174347_A_C | 199 | F>C | No | 1000Genomes | |
| ENSVATH00228629 | 199 | F>L | No | 1000Genomes | |
| ENSVATH01786905 | 214 | V>I | No | 1000Genomes | |
| ENSVATH13090598 | 220 | M>I | No | 1000Genomes | |
| ENSVATH13090596 | 230 | I>M | No | 1000Genomes | |
| ENSVATH01786903 | 233 | T>S | No | 1000Genomes | |
| tmp_2_6174127_A_T | 241 | D>E | No | 1000Genomes | |
| ENSVATH00228628 | 244 | N>K | No | 1000Genomes | |
| ENSVATH05460813 | 245 | P>L | No | 1000Genomes | |
| ENSVATH05460812 | 255 | I>V | No | 1000Genomes | |
| ENSVATH01786901 | 259 | G>V | No | 1000Genomes | |
| tmp_2_6174069_C_T | 261 | E>K | No | 1000Genomes | |
| tmp_2_6174062_A_C | 263 | L>W | No | 1000Genomes | |
| ENSVATH05460809 | 268 | N>I | No | 1000Genomes | |
| tmp_2_6174046_A_C | 268 | N>K | No | 1000Genomes | |
| tmp_2_6174036_A_C | 272 | S>A | No | 1000Genomes | |
| ENSVATH14447532 | 280 | L>I | No | 1000Genomes | |
| ENSVATH01786900 | 281 | F>Y | No | 1000Genomes | |
| ENSVATH05460808 | 284 | E>Q | No | 1000Genomes | |
| ENSVATH05460806 | 286 | Q>K | No | 1000Genomes | |
| ENSVATH01786899 | 287 | Q>K | No | 1000Genomes | |
| ENSVATH01786898 | 290 | V>A | No | 1000Genomes | |
| ENSVATH13090583 | 291 | N>H | No | 1000Genomes | |
| ENSVATH07863172 | 295 | E>Q | No | 1000Genomes | |
| ENSVATH07863170 | 299 | L>V | No | 1000Genomes | |
| ENSVATH14447531 | 302 | G>D | No | 1000Genomes | |
| ENSVATH14447531 | 302 | G>V | No | 1000Genomes | |
| ENSVATH05460804 | 303 | V>D | No | 1000Genomes | |
| ENSVATH09456744 | 303 | V>I | No | 1000Genomes | |
| ENSVATH01786897 | 309 | T>I | No | 1000Genomes | |
| tmp_2_6173918_C_A | 311 | W>L | No | 1000Genomes | |
| ENSVATH13090582 | 313 | F>L | No | 1000Genomes | |
| ENSVATH05460803 | 315 | A>S | No | 1000Genomes | |
| ENSVATH01786896 | 316 | R>G | No | 1000Genomes | |
| ENSVATH01786895 | 316 | R>K | No | 1000Genomes | |
| tmp_2_6173894_G_A | 319 | S>F | No | 1000Genomes | |
| ENSVATH05460802 | 322 | A>G | No | 1000Genomes | |
| ENSVATH01786893 | 327 | E>G | No | 1000Genomes | |
| ENSVATH05460801 | 339 | P>L | No | 1000Genomes | |
| ENSVATH07863167 | 339 | P>S | No | 1000Genomes | |
| ENSVATH13090580 | 351 | E>D | No | 1000Genomes | |
| ENSVATH05460798 | 353 | F>V | No | 1000Genomes | |
| ENSVATH13090579 | 356 | I>V | No | 1000Genomes | |
| ENSVATH01786887 | 359 | P>R | No | 1000Genomes | |
| ENSVATH07863166 | 362 | D>E | No | 1000Genomes | |
| ENSVATH01786886 | 365 | T>M | No | 1000Genomes | |
| ENSVATH01786886 | 365 | T>R | No | 1000Genomes | |
| tmp_2_6173749_T_A | 367 | E>D | No | 1000Genomes | |
| ENSVATH13090558 | 371 | I>V | No | 1000Genomes | |
| ENSVATH01786862 | 373 | K>N | No | 1000Genomes | |
| ENSVATH14447528 | 379 | Q>P | No | 1000Genomes | |
| tmp_2_6173516_T_G | 381 | S>R | No | 1000Genomes | |
| tmp_2_6173506_C_T | 384 | S>N | No | 1000Genomes | |
| tmp_2_6173486_C_G | 391 | V>L | No | 1000Genomes | |
| ENSVATH01786857 | 396 | S>T | No | 1000Genomes | |
| ENSVATH13090557 | 406 | D>Y | No | 1000Genomes | |
| ENSVATH13090555 | 412 | R>G | No | 1000Genomes | |
| ENSVATH05460782 | 415 | S>A | No | 1000Genomes | |
| ENSVATH01786850 | 420 | L>S | No | 1000Genomes | |
| tmp_2_6173321_C_A | 421 | S>I | No | 1000Genomes | |
| ENSVATH01786849 | 424 | T>I | No | 1000Genomes | |
| ENSVATH01786847 | 428 | S>* | No | 1000Genomes | |
| ENSVATH01786848 | 428 | S>P | No | 1000Genomes | |
| ENSVATH01786848 | 428 | S>T | No | 1000Genomes | |
| ENSVATH13090529 | 429 | P>A | No | 1000Genomes | |
| ENSVATH07863159 | 431 | I>L | No | 1000Genomes | |
| tmp_2_6173289_G_T | 432 | Q>K | No | 1000Genomes | |
| ENSVATH07863158 | 438 | R>Q | No | 1000Genomes | |
| ENSVATH07863157 | 439 | E>K | No | 1000Genomes | |
| tmp_2_6173176_G_C | 443 | S>* | No | 1000Genomes | |
| tmp_2_6173137_G_T | 456 | A>D | No | 1000Genomes | |
| ENSVATH01786828 | 457 | T>N | No | 1000Genomes | |
| ENSVATH01786825 | 460 | P>S | No | 1000Genomes | |
| ENSVATH01786817 | 465 | H>N | No | 1000Genomes | |
| ENSVATH13090503 | 478 | A>T | No | 1000Genomes | |
| ENSVATH01786811 | 480 | Q>H | No | 1000Genomes | |
| ENSVATH01786810 | 483 | E>K | No | 1000Genomes | |
| ENSVATH01786809 | 483 | E>V | No | 1000Genomes | |
| ENSVATH05460770 | 484 | K>N | No | 1000Genomes | |
| ENSVATH01786808 | 486 | D>E | No | 1000Genomes | |
| ENSVATH01786807 | 490 | L>I | No | 1000Genomes | |
| ENSVATH01786806 | 491 | F>Y | No | 1000Genomes | |
| ENSVATH14447472 | 492 | V>I | No | 1000Genomes | |
| ENSVATH05460762 | 497 | T>K | No | 1000Genomes | |
| ENSVATH05460761 | 498 | R>P | No | 1000Genomes | |
| ENSVATH05460761 | 498 | R>Q | No | 1000Genomes | |
| ENSVATH00228625 | 499 | R>H | No | 1000Genomes | |
| ENSVATH05460759 | 500 | G>E | No | 1000Genomes | |
| tmp_2_6172846_C_T | 500 | G>R | No | 1000Genomes | |
| ENSVATH05460758 | 501 | K>Q | No | 1000Genomes | |
| tmp_2_6172842_T_G | 501 | K>T | No | 1000Genomes | |
| tmp_2_6172837_G_T | 503 | Q>K | No | 1000Genomes | |
| ENSVATH05460756 | 503 | Q>R | No | 1000Genomes | |
| ENSVATH07863149 | 505 | K>R | No | 1000Genomes | |
| tmp_2_6172788_G_A | 519 | A>V | No | 1000Genomes | |
| ENSVATH01786793 | 520 | V>A | No | 1000Genomes | |
| tmp_2_6172783_T_A | 521 | T>S | No | 1000Genomes | |
| ENSVATH14447470 | 524 | V>M | No | 1000Genomes | |
| ENSVATH07863144 | 530 | I>V | No | 1000Genomes | |
| ENSVATH01786759 | 555 | Y>F | No | 1000Genomes | |
| tmp_2_6172394_A_G | 581 | F>S | No | 1000Genomes | |
| tmp_2_6172338_A_T | 600 | Y>N | No | 1000Genomes | |
| tmp_2_6172227_C_T | 608 | E>K | No | 1000Genomes | |
| ENSVATH13090450 | 612 | R>K | No | 1000Genomes | |
| ENSVATH00228624 | 629 | I>N | No | 1000Genomes | |
| ENSVATH01786744 | 634 | I>V | No | 1000Genomes | |
| ENSVATH01786742 | 645 | E>D | No | 1000Genomes | |
| ENSVATH13090448 | 651 | R>H | No | 1000Genomes | |
| ENSVATH13090447 | 653 | G>C | No | 1000Genomes | |
| tmp_2_6171993_A_G | 654 | S>P | No | 1000Genomes | |
| ENSVATH01786728 | 659 | S>P | No | 1000Genomes | |
| ENSVATH05460723 | 660 | S>G | No | 1000Genomes | |
| ENSVATH14447262 | 660 | S>N | No | 1000Genomes | |
| ENSVATH01786727 | 662 | L>F | No | 1000Genomes | |
| ENSVATH01786727 | 662 | L>I | No | 1000Genomes | |
| ENSVATH01786726 | 663 | K>E | No | 1000Genomes | |
| ENSVATH13090446 | 663 | K>N | No | 1000Genomes | |
| ENSVATH01786724 | 669 | A>G | No | 1000Genomes | |
| tmp_2_6171805_C_T | 678 | G>R | No | 1000Genomes | |
| ENSVATH01786709 | 680 | Q>K | No | 1000Genomes | |
| ENSVATH01786708 | 680 | Q>P | No | 1000Genomes | |
| ENSVATH00228623 | 683 | M>I | No | 1000Genomes | |
| tmp_2_6171751_C_T | 696 | G>S | No | 1000Genomes | |
| tmp_2_6171689_A_C | 716 | S>R | No | 1000Genomes | |
| ENSVATH01786700 | 718 | A>G | No | 1000Genomes | |
| ENSVATH01786701 | 718 | A>T | No | 1000Genomes | |
| ENSVATH05460713 | 723 | N>T | No | 1000Genomes | |
| tmp_2_6171661_C_T | 726 | G>R | No | 1000Genomes | |
| ENSVATH07863109 | 737 | L>Q | No | 1000Genomes | |
| ENSVATH01786681 | 753 | V>L | No | 1000Genomes | |
| ENSVATH01786679 | 757 | S>I | No | 1000Genomes | |
| tmp_2_6171464_G_A | 759 | T>I | No | 1000Genomes | |
| ENSVATH13090401 | 764 | I>M | No | 1000Genomes | |
| tmp_2_6171449_A_C | 764 | I>S | No | 1000Genomes | |
| ENSVATH13090400 | 769 | D>N | No | 1000Genomes | |
| ENSVATH13090399 | 771 | S>C | No | 1000Genomes | |
| ENSVATH13090398 | 772 | Y>F | No | 1000Genomes | |
| ENSVATH13090396 | 783 | N>H | No | 1000Genomes | |
| tmp_2_6171345_C_T | 799 | D>N | No | 1000Genomes | |
| ENSVATH01786670 | 800 | S>A | No | 1000Genomes | |
| ENSVATH01786670 | 800 | S>T | No | 1000Genomes | |
| ENSVATH13090384 | 820 | Q>H | No | 1000Genomes | |
| tmp_2_6171281_T_C,A | 820 | Q>L | No | 1000Genomes | |
| tmp_2_6171281_T_C,A | 820 | Q>R | No | 1000Genomes | |
| ENSVATH05460705 | 839 | N>K | No | 1000Genomes | |
| ENSVATH07863100 | 843 | I>K | No | 1000Genomes | |
| ENSVATH05460704 | 846 | Q>H | No | 1000Genomes | |
| ENSVATH05460703 | 848 | Q>P | No | 1000Genomes | |
| ENSVATH01786664 | 849 | N>I | No | 1000Genomes | |
| ENSVATH07863099 | 854 | L>S | No | 1000Genomes | |
| tmp_2_6171174_G_C | 856 | H>D | No | 1000Genomes | |
| ENSVATH09456692 | 858 | V>L | No | 1000Genomes | |
| tmp_2_6171159_T_C | 861 | I>V | No | 1000Genomes | |
| ENSVATH13090382 | 868 | R>H | No | 1000Genomes |
No associated diseases with Q9ZQR3
3 regional properties for Q9ZQR3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Heat shock protein 70, conserved site | 11 - 18 | IPR018181-1 |
| conserved_site | Heat shock protein 70, conserved site | 199 - 212 | IPR018181-2 |
| conserved_site | Heat shock protein 70, conserved site | 336 - 350 | IPR018181-3 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
36 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q91044 | NTRK3 | NT-3 growth factor receptor | Gallus gallus (Chicken) | SS |
| Q91987 | NTRK2 | BDNF/NT-3 growth factors receptor | Gallus gallus (Chicken) | SS |
| Q91009 | NTRK1 | High affinity nerve growth factor receptor | Gallus gallus (Chicken) | SS |
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| P20594 | NPR2 | Atrial natriuretic peptide receptor 2 | Homo sapiens (Human) | PR |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P24786 | NTRK3 | NT-3 growth factor receptor | Sus scrofa (Pig) | SS |
| P35739 | Ntrk1 | High affinity nerve growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q03351 | Ntrk3 | NT-3 growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q63604 | Ntrk2 | BDNF/NT-3 growth factors receptor | Rattus norvegicus (Rat) | SS |
| Q19238 | F09A5.2 | Putative tyrosine-protein kinase F09A5.2 | Caenorhabditis elegans | SS |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGG3 | At1g51820 | Probable LRR receptor-like serine/threonine-protein kinase At1g51820 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGW2 | PAM74 | Probable LRR receptor-like serine/threonine-protein kinase PAM74 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FN93 | At5g59680 | Probable LRR receptor-like serine/threonine-protein kinase At5g59680 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FN94 | At5g59670 | Receptor-like protein kinase At5g59670 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LIG2 | At3g21340 | Receptor-like protein kinase At3g21340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SNA3 | At3g46340 | Putative receptor-like protein kinase At3g46340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LPZ9 | SD113 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P43298 | TMK1 | Receptor protein kinase TMK1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGI2 | At1g67720 | Probable LRR receptor-like serine/threonine-protein kinase At1g67720 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2S4 | LECRKS4 | L-type lectin-domain containing receptor kinase S.4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWZ5 | SD25 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64556 | At2g19230 | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g19230 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SXB4 | At1g11300 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q84JQ4 | PRK2 | Pollen receptor-like kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q93ZS4 | NIK3 | Protein NSP-INTERACTING KINASE 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94AG2 | SERK1 | Somatic embryogenesis receptor kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64477 | At2g19130 | G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9XIC7 | SERK2 | Somatic embryogenesis receptor kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| METRNKFMLL | ACATFSIMSL | VKSQNQQGFI | SLDCGLPSKE | SYIEPSSNLT | FISDVNFIRG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GKTGNIQNNS | RTNFIFKPFK | VLRYFPDGIR | NCYSLSVKQG | TKYLIRTLFY | YGNYDGLNTS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PRFDLFLGPN | IWTSVDVLIA | DVGDGVVEEI | VHVTRSNILD | ICLVKTGTST | PMISAIELRP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LRYDTYTART | GSLKSMAHFY | FTNSDEAIRY | PEDVYDRVWM | PYSQPEWTQI | NTTRNVSGFS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DGYNPPQGVI | QTASIPTNGS | EPLTFTWNLE | SSDDETYAYL | FFAEIQQLKV | NETREFKILA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NGVDYIDYTP | WKFEARTLSN | PAPLKCEGGV | CRVQLSKTPK | STLPPLMNAI | EIFSVIQFPQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SDTNTDEVIA | IKKIQSTYQL | SRISWQGDPC | VPKQFSWMGV | SCNVIDISTP | PRIISLDLSL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SGLTGVISPS | IQNLTMLREL | DLSNNNLTGE | VPEFLATIKP | LLVIHLRGNN | LRGSVPQALQ |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DREKNDGLKL | FVDPNITRRG | KHQPKSWLVA | IVASISCVAV | TIIVLVLIFI | FRRRKSSTRK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| VIRPSLEMKN | RRFKYSEVKE | MTNNFEVVLG | KGGFGVVYHG | FLNNEQVAVK | VLSQSSTQGY |
| 610 | 620 | 630 | 640 | 650 | 660 |
| KEFKTEVELL | LRVHHVNLVS | LVGYCDEGID | LALIYEFMEN | GNLKEHLSGK | RGGSVLNWSS |
| 670 | 680 | 690 | 700 | 710 | 720 |
| RLKIAIESAL | GIEYLHIGCQ | PPMVHRDVKS | TNILLGLRFE | AKLADFGLSR | SFLVGSQAHV |
| 730 | 740 | 750 | 760 | 770 | 780 |
| STNVAGTLGY | LDPEYYLKNW | LTEKSDVYSF | GIVLLESITG | QPVIEQSRDK | SYIVEWAKSM |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LANGDIESIM | DPNLHQDYDS | SSSWKALELA | MLCINPSSTQ | RPNMTRVAHE | LNECLEIYNL |
| 850 | 860 | ||||
| TKIRSQDQNS | SKSLGHTVTF | ISDIPSAR |