Q9Z1K6
Gene name |
Arih2 (Ari2, Triad1, Uip48) |
Protein name |
E3 ubiquitin-protein ligase ARIH2 |
Names |
ARI-2, Protein ariadne-2 homolog, RING-type E3 ubiquitin transferase ARIH2, Triad1 protein, UbcM4-interacting protein 48 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:23807 |
EC number |
2.3.2.31: Aminoacyltransferases |
Protein Class |
|
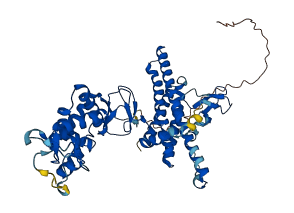
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
299-339 (RING2 domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Duda DM et al. (2013) "Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism", Structure (London, England : 1993), 21, 1030-41
- Kelsall IR et al. (2013) "TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes", The EMBO journal, 32, 2848-60
Autoinhibited structure

Activated structure

1 structures for Q9Z1K6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9Z1K6-F1 | Predicted | AlphaFoldDB |
2 variants for Q9Z1K6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs46596494 | 42 | A>V | No | Ensembl | |
| rs235516447 | 386 | E>Q | No | Ensembl |
No associated diseases with Q9Z1K6
8 regional properties for Q9Z1K6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Zinc finger, RING-type | 138 - 189 | IPR001841-1 |
| domain | Zinc finger, RING-type | 299 - 339 | IPR001841-2 |
| domain | IBR domain | 207 - 269 | IPR002867-1 |
| domain | IBR domain | 277 - 339 | IPR002867-2 |
| domain | TRIAD supradomain | 134 - 343 | IPR044066 |
| domain | Ariadne domain | 352 - 469 | IPR045840 |
| domain | E3 ubiquitin-protein ligase ARIH2, BRcat domain | 216 - 287 | IPR047555 |
| domain | E3 ubiquitin-protein ligase TRIAD1, Rcat domain | 291 - 346 | IPR047556 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.31 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| Cul5-RING ubiquitin ligase complex | A ubiquitin ligase complex in which a cullin from the Cul5 subfamily and a RING domain protein form the catalytic core; substrate specificity is conferred by an elongin-BC adaptor and a SOCS/BC box protein. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| ubiquitin ligase complex | A protein complex that includes a ubiquitin-protein ligase and enables ubiquitin protein ligase activity. The complex also contains other proteins that may confer substrate specificity on the complex. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| metal ion binding | Binding to a metal ion. |
| ubiquitin conjugating enzyme binding | Binding to a ubiquitin conjugating enzyme, any of the E2 proteins. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| developmental cell growth | The growth of a cell, where growth contributes to the progression of the cell over time from one condition to another. |
| hematopoietic stem cell proliferation | The expansion of a hematopoietic stem cell population by cell division. A hematopoietic stem cell is a stem cell from which all cells of the lymphoid and myeloid lineages develop. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| protein K48-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 48 of the ubiquitin monomers, is added to a protein. K48-linked ubiquitination targets the substrate protein for degradation. |
| protein K63-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 63 of the ubiquitin monomers, is added to a protein. K63-linked ubiquitination does not target the substrate protein for degradation, but is involved in several pathways, notably as a signal to promote error-free DNA postreplication repair. |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of a ubiquitin group, or multiple ubiquitin groups, to the protein. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A2VEA3 | ARIH1 | E3 ubiquitin-protein ligase ARIH1 | Bos taurus (Bovine) | SS |
| O76924 | ari-2 | Potential E3 ubiquitin-protein ligase ariadne-2 | Drosophila melanogaster (Fruit fly) | SS |
| Q94981 | ari-1 | E3 ubiquitin-protein ligase ariadne-1 | Drosophila melanogaster (Fruit fly) | SS |
| Q9NWF9 | RNF216 | E3 ubiquitin-protein ligase RNF216 | Homo sapiens (Human) | PR |
| O95376 | ARIH2 | E3 ubiquitin-protein ligase ARIH2 | Homo sapiens (Human) | EV |
| Q9Y4X5 | ARIH1 | E3 ubiquitin-protein ligase ARIH1 | Homo sapiens (Human) | EV |
| Q9WVS6 | Prkn | E3 ubiquitin-protein ligase parkin | Mus musculus (Mouse) | EV SS |
| Q9Z1K5 | Arih1 | E3 ubiquitin-protein ligase ARIH1 | Mus musculus (Mouse) | SS |
| Q22431 | ari-2 | Potential E3 ubiquitin-protein ligase ariadne-2 | Caenorhabditis elegans | SS |
| O01965 | ari-1.1 | E3 ubiquitin-protein ligase ari-1.1 | Caenorhabditis elegans | SS |
| B1H1E4 | arih1 | E3 ubiquitin-protein ligase arih1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q6PFJ9 | arih1 | E3 ubiquitin-protein ligase arih1 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q6NW85 | arih1l | E3 ubiquitin-protein ligase arih1l | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSVDMNSQGS | DSNEEDYDPN | CEEEEEEEED | PGDIEDYYVG | VASDVEQQGA | DAFDPEEYQF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TCLTYKESEG | ALHEHMTSLA | SVLKVSHSVA | KLILVNFHWQ | VSEILDRYRS | NSAQLLVEAR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VQPNPSKHVP | TAHPPHHCAV | CMQFVRKENL | LSLACQHQFC | RSCWEQHCSV | LVKDGVGVGI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SCMAQDCPLR | TPEDFVFPLL | PNEELRDKYR | RYLFRDYVES | HFQLQLCPGA | DCPMVIRVQE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PRARRVQCNR | CSEVFCFKCR | QMYHAPTDCA | TIRKWLTKCA | DDSETANYIS | AHTKDCPKCN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ICIEKNGGCN | HMQCSKCKHD | FCWMCLGDWK | THGSEYYECS | RYKENPDIVN | QSQQAQAREA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LKKYLFYFER | WENHNKSLQL | EAQTYERIHE | KIQERVMNNL | GTWIDWQYLQ | NAAKLLAKCR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| YTLQYTYPYA | YYMESGPRKK | LFEYQQAQLE | AEIENLSWKV | ERADSYDRGD | LENQMHIAEQ |
| 490 | |||||
| RRRTLLKDFH | DT |