Q9Y689
Gene name |
ARL5A (ARFLP5, ARL5) |
Protein name |
ADP-ribosylation factor-like protein 5A |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:26225 |
EC number |
|
Protein Class |
|
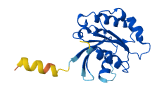
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
15-137 (Small GTP-binding protein domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for Q9Y689
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1Z6Y | X-ray | 240 A | A/B | 1-179 | PDB |
| 1ZJ6 | X-ray | 200 A | A | 1-179 | PDB |
| 2H16 | X-ray | 200 A | A/B/C/D | 14-177 | PDB |
| 2H17 | X-ray | 170 A | A | 15-176 | PDB |
| AF-Q9Y689-F1 | Predicted | AlphaFoldDB |
174 variants for Q9Y689
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| COSM3567930 | 1 | M>? | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2099833348 | 2 | G>R | No | TOPMed | |
| rs1215342829 | 4 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1215342829 | 4 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1364551535 | 4 | L>P | No | gnomAD | |
| rs1215342829 | 4 | L>V | No |
TOPMed gnomAD |
|
| rs2099833347 | 5 | F>L | No | TOPMed | |
| rs749519458 | 6 | T>A | No |
ExAC gnomAD |
|
| rs1450803725 | 7 | R>G | No |
TOPMed gnomAD |
|
| rs1328499301 | 8 | I>M | No |
TOPMed gnomAD |
|
| rs778324868 | 8 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs567730459 | 10 | R>G | No |
1000Genomes ExAC gnomAD |
|
| rs2099833344 | 10 | R>K | No | Ensembl | |
| rs1404448310 | 10 | R>S | No | gnomAD | |
| rs781515447 | 12 | F>V | No |
ExAC gnomAD |
|
| rs1458042804 | 13 | N>D | No |
TOPMed gnomAD |
|
| rs754523394 | 13 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751016607 | 15 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs751016607 | 15 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs765866459 | 15 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs933836843 | 18 | K>E | No | TOPMed | |
| rs1426993825 | 18 | K>R | No | TOPMed | |
| rs757088388 | 19 | V>F | No |
ExAC gnomAD |
|
| rs922494082 | 20 | I>T | No | TOPMed | |
| rs1316889209 | 21 | I>L | No |
TOPMed gnomAD |
|
| rs1316889209 | 21 | I>V | No |
TOPMed gnomAD |
|
| rs2099831335 | 22 | V>A | No | TOPMed | |
| rs1308335084 | 22 | V>I | No |
TOPMed gnomAD |
|
| rs1007352443 | 25 | D>E | No | gnomAD | |
| rs543220434 | 26 | N>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1286810408 | 27 | A>P | No |
TOPMed gnomAD |
|
| rs1286810408 | 27 | A>S | No |
TOPMed gnomAD |
|
| rs752660825 | 28 | G>R | No |
ExAC gnomAD |
|
| rs1451353692 | 30 | T>S | No | gnomAD | |
| rs1363210155 | 31 | T>N | No | gnomAD | |
| rs2099831332 | 35 | Q>E | No | Ensembl | |
| TCGA novel | 36 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766588581 | 36 | F>S | No |
ExAC gnomAD |
|
| rs1191231176 | 37 | S>F | No | gnomAD | |
| rs1262819845 | 38 | M>I | No | gnomAD | |
| rs2099831215 | 38 | M>T | No |
TOPMed gnomAD |
|
| rs760103873 | 38 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs2099831213 | 39 | N>K | No |
TOPMed gnomAD |
|
| rs763142365 | 40 | E>D | No |
ExAC gnomAD |
|
| rs750528869 | 42 | V>A | No |
ExAC gnomAD |
|
| rs765357610 | 43 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1273101156 | 43 | H>Y | No |
TOPMed gnomAD |
|
| rs1356360996 | 44 | T>I | No |
TOPMed gnomAD |
|
| rs2099831211 | 44 | T>S | No |
TOPMed gnomAD |
|
| rs2151294523 | 45 | S>F | No | Ensembl | |
| rs2099831210 | 45 | S>T | No | TOPMed | |
| rs1180940820 | 47 | T>K | No |
TOPMed gnomAD |
|
| rs1475939197 | 48 | I>T | No | TOPMed | |
| rs1325660594 | 48 | I>V | No | gnomAD | |
| rs2099831209 | 52 | V>I | No | gnomAD | |
| rs760960104 | 55 | I>L | No |
ExAC gnomAD |
|
| rs199870857 | 55 | I>M | No | Ensembl | |
| rs1006651984 | 55 | I>R | No |
TOPMed gnomAD |
|
| rs1006651984 | 55 | I>T | No |
TOPMed gnomAD |
|
| rs2099831207 | 56 | V>A | No |
TOPMed gnomAD |
|
| rs2099831207 | 56 | V>E | No |
TOPMed gnomAD |
|
| rs775988971 | 56 | V>M | No |
ExAC gnomAD |
|
| rs746379591 | 58 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs2099831206 | 59 | N>K | No | TOPMed | |
| rs2099831205 | 60 | T>A | No | Ensembl | |
| rs1461628035 | 60 | T>K | No | gnomAD | |
| rs778325948 | 61 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs778325948 | 61 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1452947636 | 61 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2099831204 | 64 | M>I | No | TOPMed | |
| rs777125453 | 64 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1268776848 | 67 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2099831203 | 71 | E>A | No | Ensembl | |
| rs1189090837 | 71 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| COSM716330 | 71 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1338396146 | 71 | E>Q | No |
TOPMed gnomAD |
|
| rs1311364380 | 72 | S>C | No | TOPMed | |
| rs755979325 | 72 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1230416518 | 74 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs747897247 | 74 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs747897247 | 74 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs781125420 | 75 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs2099831202 | 76 | S>F | No | Ensembl | |
| rs2151294486 | 77 | W>C | No | Ensembl | |
| rs755036544 | 78 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs142801727 | 79 | T>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs2099831201 | 79 | T>I | No |
TOPMed gnomAD |
|
| rs2099831199 | 80 | Y>* | No | TOPMed | |
| rs757315405 | 81 | Y>C | No |
ExAC gnomAD |
|
| rs757315405 | 81 | Y>F | No |
ExAC gnomAD |
|
| rs754007177 | 82 | T>A | No |
ExAC gnomAD |
|
| rs2099831198 | 82 | T>I | No | gnomAD | |
| rs2099831196 | 83 | N>D | No | Ensembl | |
| rs1433248227 | 83 | N>K | No | gnomAD | |
| rs1394811887 | 85 | E>K | No | gnomAD | |
| rs1174837796 | 85 | E>V | No | gnomAD | |
| rs764155367 | 88 | I>L | No |
ExAC gnomAD |
|
| rs764155367 | 88 | I>V | No |
ExAC gnomAD |
|
| rs756366503 | 89 | V>A | No |
ExAC gnomAD |
|
| rs2099830963 | 90 | V>G | No | TOPMed | |
| rs1231397393 | 90 | V>I | No |
TOPMed gnomAD |
|
| rs773494531 | 94 | T>R | No | Ensembl | |
|
rs2099830960 COSM441261 |
96 | R>G | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs952948687 | 98 | R>G | No | Ensembl | |
| rs767891575 | 98 | R>K | No |
ExAC gnomAD |
|
| rs952948687 | 98 | R>W | No | Ensembl | |
| rs2099830958 | 101 | V>I | No | TOPMed | |
| rs2099830958 | 101 | V>L | No | TOPMed | |
| rs1282477833 | 102 | T>A | No | TOPMed | |
| rs759905887 | 103 | R>G | No |
ExAC gnomAD |
|
| rs2099830957 | 103 | R>T | No | gnomAD | |
| rs774726314 | 104 | E>* | No |
ExAC gnomAD |
|
| rs774726314 | 104 | E>K | No |
ExAC gnomAD |
|
| rs748250229 | 107 | Y>* | No |
ExAC gnomAD |
|
| rs2099830955 | 107 | Y>C | No | Ensembl | |
| rs763359699 | 107 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs532438026 | 108 | K>I | No |
1000Genomes TOPMed gnomAD |
|
| rs1484944071 | 109 | M>I | No | Ensembl | |
| rs1231495089 | 110 | L>F | No |
TOPMed gnomAD |
|
| TCGA novel | 111 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs141663338 | 111 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1169194447 | 112 | H>L | No | gnomAD | |
| rs1169194447 | 112 | H>R | No | gnomAD | |
| rs1559819928 | 112 | H>Y | No | Ensembl | |
| rs143420605 | 113 | E>G | No |
ESP TOPMed gnomAD |
|
| rs2151292187 | 114 | D>Y | No | Ensembl | |
| rs748170373 | 116 | R>T | No |
ExAC gnomAD |
|
| rs781119879 | 122 | I>S | No |
ExAC gnomAD |
|
| rs1430171563 | 122 | I>V | No | gnomAD | |
| rs181841961 | 125 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 127 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751702231 | 130 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1425751740 | 133 | M>V | No | gnomAD | |
| rs2099830175 | 135 | V>I | No | gnomAD | |
| COSM4085613 | 136 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 137 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780521220 | 137 | E>Q | No | ExAC | |
| rs750966947 | 138 | I>V | No |
ExAC gnomAD |
|
| rs1351336055 | 139 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs765778672 | 140 | Q>E | No |
ExAC gnomAD |
|
| rs1299482670 | 141 | F>C | No |
TOPMed gnomAD |
|
| rs753452924 | 144 | L>V | No |
ExAC gnomAD |
|
| rs2099830173 | 147 | I>F | No |
TOPMed gnomAD |
|
| rs760425089 | 147 | I>T | No | ExAC | |
| rs2099830173 | 147 | I>V | No |
TOPMed gnomAD |
|
| rs1304030720 | 149 | D>V | No | gnomAD | |
| rs563274203 | 149 | D>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2099830172 | 151 | Q>H | No | Ensembl | |
| rs1370457140 | 153 | H>R | No | gnomAD | |
| TCGA novel | 153 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1578371960 | 154 | I>M | No | Ensembl | |
| rs977941930 | 156 | A>S | No |
TOPMed gnomAD |
|
| rs977941930 | 156 | A>T | No |
TOPMed gnomAD |
|
| rs759259880 | 156 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1207699366 | 157 | C>* | No | Ensembl | |
| rs2099830171 | 157 | C>F | No | gnomAD | |
| TCGA novel | 157 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2099830170 | 159 | A>T | No | gnomAD | |
| rs1465357376 | 163 | E>G | No |
TOPMed gnomAD |
|
| rs770913765 | 163 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1464876435 | 165 | L>V | No | gnomAD | |
| rs151249497 | 168 | G>R | No |
ESP ExAC gnomAD |
|
| rs746999786 | 171 | W>* | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 172 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2151290927 | 172 | M>T | No | Ensembl | |
| rs1559816950 | 173 | M>V | No | TOPMed | |
| rs772485630 | 175 | R>* | No |
ExAC gnomAD |
|
| rs138455197 | 175 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs202064303 | 176 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2151290917 | 176 | L>P | No | Ensembl | |
| rs189649167 | 177 | K>E | No |
1000Genomes gnomAD |
|
| rs1321074798 | 178 | I>M | No | gnomAD | |
| rs1421926619 | 178 | I>S | No |
TOPMed gnomAD |
|
| COSM5631876 | 179 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
No associated diseases with Q9Y689
1 regional properties for Q9Y689
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 15 - 143 | IPR005225 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| trans-Golgi network | The network of interconnected tubular and cisternal structures located within the Golgi apparatus on the side distal to the endoplasmic reticulum, from which secretory vesicles emerge. The trans-Golgi network is important in the later stages of protein secretion where it is thought to play a key role in the sorting and targeting of secreted proteins to the correct destination. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| protein localization to Golgi membrane | A process in which a protein is transported to, or maintained in, a location within a Golgi membrane. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
48 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P84081 | ARF2 | ADP-ribosylation factor 2 | Bos taurus (Bovine) | SS |
| P84080 | ARF1 | ADP-ribosylation factor 1 | Bos taurus (Bovine) | SS |
| Q3SZF2 | ARF4 | ADP-ribosylation factor 4 | Bos taurus (Bovine) | SS |
| Q0VC18 | ARL4D | ADP-ribosylation factor-like protein 4D | Bos taurus (Bovine) | PR |
| Q2KJ96 | ARL5A | ADP-ribosylation factor-like protein 5A | Bos taurus (Bovine) | SS |
| P49702 | ARF5 | ADP-ribosylation factor 5 | Gallus gallus (Chicken) | SS |
| P40945 | Arf4 | ADP ribosylation factor 4 | Drosophila melanogaster (Fruit fly) | SS |
| P61209 | Arf1 | ADP-ribosylation factor 1 | Drosophila melanogaster (Fruit fly) | SS |
| Q8N4G2 | ARL14 | ADP-ribosylation factor-like protein 14 | Homo sapiens (Human) | SS |
| Q969Q4 | ARL11 | ADP-ribosylation factor-like protein 11 | Homo sapiens (Human) | PR |
| Q8IVW1 | ARL17A | ADP-ribosylation factor-like protein 17 | Homo sapiens (Human) | SS |
| P49703 | ARL4D | ADP-ribosylation factor-like protein 4D | Homo sapiens (Human) | PR |
| P62330 | ARF6 | ADP-ribosylation factor 6 | Homo sapiens (Human) | EV |
| Q9H0F7 | ARL6 | ADP-ribosylation factor-like protein 6 | Homo sapiens (Human) | SS |
| P40616 | ARL1 | ADP-ribosylation factor-like protein 1 | Homo sapiens (Human) | SS |
| P18085 | ARF4 | ADP-ribosylation factor 4 | Homo sapiens (Human) | EV |
| P84085 | ARF5 | ADP-ribosylation factor 5 | Homo sapiens (Human) | EV |
| P61204 | ARF3 | ADP-ribosylation factor 3 | Homo sapiens (Human) | EV |
| P84077 | ARF1 | ADP-ribosylation factor 1 | Homo sapiens (Human) | EV |
| A6NH57 | ARL5C | Putative ADP-ribosylation factor-like protein 5C | Homo sapiens (Human) | SS |
| Q96KC2 | ARL5B | ADP-ribosylation factor-like protein 5B | Homo sapiens (Human) | SS |
| P49076 | ARF1 | ADP-ribosylation factor | Zea mays (Maize) | SS |
| P84084 | Arf5 | ADP-ribosylation factor 5 | Mus musculus (Mouse) | SS |
| P61750 | Arf4 | ADP-ribosylation factor 4 | Mus musculus (Mouse) | SS |
| Q99PE9 | Arl4d | ADP-ribosylation factor-like protein 4D | Mus musculus (Mouse) | PR |
| P61205 | Arf3 | ADP-ribosylation factor 3 | Mus musculus (Mouse) | SS |
| P84078 | Arf1 | ADP-ribosylation factor 1 | Mus musculus (Mouse) | SS |
| Q8BSL7 | Arf2 | ADP-ribosylation factor 2 | Mus musculus (Mouse) | SS |
| Q6P068 | Arl5c | ADP-ribosylation factor-like protein 5C | Mus musculus (Mouse) | SS |
| Q9D4P0 | Arl5b | ADP-ribosylation factor-like protein 5B | Mus musculus (Mouse) | SS |
| Q80ZU0 | Arl5a | ADP-ribosylation factor-like protein 5A | Mus musculus (Mouse) | SS |
| P51824 | ADP-ribosylation factor 1 | Solanum tuberosum (Potato) | SS | |
| P84082 | Arf2 | ADP-ribosylation factor 2 | Rattus norvegicus (Rat) | SS |
| P84083 | Arf5 | ADP-ribosylation factor 5 | Rattus norvegicus (Rat) | SS |
| P36407 | Trim23 | E3 ubiquitin-protein ligase TRIM23 | Rattus norvegicus (Rat) | SS |
| P61751 | Arf4 | ADP-ribosylation factor 4 | Rattus norvegicus (Rat) | SS |
| P61206 | Arf3 | ADP-ribosylation factor 3 | Rattus norvegicus (Rat) | SS |
| P84079 | Arf1 | ADP-ribosylation factor 1 | Rattus norvegicus (Rat) | SS |
| P51646 | Arl5a | ADP-ribosylation factor-like protein 5A | Rattus norvegicus (Rat) | SS |
| Q06396 | Os01g0813400 | ADP-ribosylation factor 1 | Oryza sativa subsp. japonica (Rice) | SS |
| P51823 | ARF | ADP-ribosylation factor 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q10943 | arf-1.2 | ADP-ribosylation factor 1-like 2 | Caenorhabditis elegans | SS |
| P34212 | arl-5 | ADP-ribosylation factor-like protein 5 | Caenorhabditis elegans | SS |
| Q9LQC8 | ARF2-A | ADP-ribosylation factor 2-A | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P0DH91 | ARF2-B | ADP-ribosylation factor 2-B | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P40940 | ARF3 | ADP-ribosylation factor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P36397 | ARF1 | ADP-ribosylation factor 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5M9P8 | arl6 | ADP-ribosylation factor-like protein 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGILFTRIWR | LFNHQEHKVI | IVGLDNAGKT | TILYQFSMNE | VVHTSPTIGS | NVEEIVINNT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RFLMWDIGGQ | ESLRSSWNTY | YTNTEFVIVV | VDSTDRERIS | VTREELYKML | AHEDLRKAGL |
| 130 | 140 | 150 | 160 | 170 | |
| LIFANKQDVK | ECMTVAEISQ | FLKLTSIKDH | QWHIQACCAL | TGEGLCQGLE | WMMSRLKIR |