Q9Y5Y7
Gene name |
LYVE1 (CRSBP1, HAR, XLKD1, UNQ230/PRO263) |
Protein name |
Lymphatic vessel endothelial hyaluronic acid receptor 1 |
Names |
LYVE-1, Cell surface retention sequence-binding protein 1, CRSBP-1, Extracellular link domain-containing protein 1, Hyaluronic acid receptor |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:10894 |
EC number |
|
Protein Class |
|
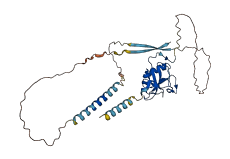
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9Y5Y7
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9Y5Y7-F1 | Predicted | AlphaFoldDB |
263 variants for Q9Y5Y7
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1199189534 CA379655381 |
2 | A>G | No |
ClinGen gnomAD |
|
|
rs762872131 CA5883807 |
6 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs79727467 CA217669661 |
9 | L>* | No |
ClinGen Ensembl |
|
|
CA217669665 rs77800052 |
9 | L>M | No |
ClinGen Ensembl |
|
|
rs76181502 CA217669656 |
10 | L>H | No |
ClinGen Ensembl |
|
|
rs761190228 CA5883804 |
11 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA5883803 rs145001787 |
12 | T>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA379655319 rs1269805849 |
12 | T>I | No |
ClinGen TOPMed |
|
| TCGA novel | 13 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA379655312 rs1463361432 |
14 | I>L | No |
ClinGen TOPMed |
|
|
rs746630267 CA5883801 |
16 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771190217 CA5883799 |
17 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA5883798 rs749679062 |
17 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA379655271 rs1160494498 |
20 | L>P | No |
ClinGen TOPMed |
|
|
rs370756319 CA5883793 |
23 | G>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA217669596 rs377222811 |
24 | S>F | No |
ClinGen ESP TOPMed |
|
|
CA217669590 rs1027875981 |
25 | L>W | No |
ClinGen Ensembl |
|
|
CA5883791 rs766644059 |
26 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758069812 COSM428323 CA5883790 |
26 | R>H | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1194262676 CA379655229 |
28 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA379655228 rs1194262676 |
28 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA379655221 rs1290108163 |
29 | E>K | No |
ClinGen TOPMed |
|
|
CA5883775 rs747105019 |
30 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA379654830 rs1323276897 |
31 | S>F | No |
ClinGen gnomAD |
|
|
rs1196188971 CA379654840 |
31 | S>P | No |
ClinGen gnomAD |
|
|
rs1338564200 CA379654817 |
32 | I>T | No |
ClinGen gnomAD |
|
|
CA379654825 rs1460206771 |
32 | I>V | No |
ClinGen TOPMed |
|
|
rs780183406 CA5883774 |
34 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA5883771 rs778739711 |
36 | C>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757068480 CA379654742 |
36 | C>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5883768 rs764143601 |
39 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753816552 CA5883769 |
39 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs760200763 CA5883767 |
41 | I>S | No |
ClinGen ExAC gnomAD |
|
|
rs760200763 CA379654710 |
41 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA217666670 rs1048193450 |
42 | T>A | No |
ClinGen Ensembl |
|
|
CA379654705 rs770932220 |
42 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs770932220 CA217666669 |
42 | T>N | No |
ClinGen TOPMed gnomAD |
|
|
CA217666645 rs370231812 |
43 | L>P | No |
ClinGen ESP TOPMed |
|
|
rs767024076 CA5883765 |
44 | V>M | No |
ClinGen ExAC |
|
|
rs1338571616 CA379654676 |
47 | K>* | No |
ClinGen TOPMed |
|
|
rs1165857871 CA379654673 |
47 | K>T | No |
ClinGen gnomAD |
|
| TCGA novel | 48 | A>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5883764 rs759199614 |
48 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA5883763 rs141249676 COSM922521 |
48 | A>V | Variant assessed as Somatic; 0.0 impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs146540416 CA5883761 |
50 | Q>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5883760 rs146540416 |
50 | Q>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA379654654 rs1364436943 |
50 | Q>H | No |
ClinGen gnomAD |
|
|
CA217666590 rs949019231 |
51 | Q>R | No |
ClinGen Ensembl |
|
|
rs375834783 CA5883759 |
55 | T>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA379654608 rs1249358871 |
57 | A>G | No |
ClinGen gnomAD |
|
|
rs1436291521 CA379654609 |
57 | A>S | No |
ClinGen gnomAD |
|
|
rs1290897920 CA379654591 |
59 | E>D | No |
ClinGen gnomAD |
|
|
rs1490499830 CA379654593 |
59 | E>G | No |
ClinGen gnomAD |
|
|
CA5883756 rs143843236 |
60 | A>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA5883757 rs143843236 |
60 | A>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs746100946 CA5883755 |
61 | C>S | No |
ClinGen ExAC gnomAD |
|
|
CA379654582 rs746100946 |
61 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs149004349 CA5883753 |
65 | G>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs753695278 CA5883752 |
65 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5883751 rs202234918 |
66 | L>P | No |
ClinGen ExAC TOPMed |
|
| TCGA novel | 69 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs79541972 CA379654518 |
70 | G>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs79541972 CA5883748 |
70 | G>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs754774378 CA5883747 |
71 | K>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA379654469 rs1373591005 |
73 | Q>K | No |
ClinGen TOPMed |
|
|
rs1364348860 CA379654436 |
75 | E>Q | No |
ClinGen gnomAD |
|
|
CA5883745 rs754510091 |
76 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765958895 CA5883744 |
77 | A>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5883743 rs765958895 |
77 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs548675012 CA5883741 |
80 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA379654333 rs1201359067 |
81 | S>N | No |
ClinGen gnomAD |
|
|
CA379654322 rs1386492796 |
81 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA5883740 rs764492703 |
82 | F>S | No |
ClinGen ExAC gnomAD |
|
|
rs946575494 CA217666501 |
84 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs946575494 CA379654264 |
84 | T>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1448655715 CA379654232 |
86 | S>N | No |
ClinGen gnomAD |
|
|
rs776009596 CA5883720 |
87 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA217666348 rs1023052438 |
87 | Y>N | No |
ClinGen TOPMed |
|
|
rs1305798671 CA379654123 |
89 | W>* | No |
ClinGen TOPMed |
|
|
rs1270170995 CA379654126 |
89 | W>* | No |
ClinGen gnomAD |
|
|
rs1305798671 CA379654121 |
89 | W>C | No |
ClinGen TOPMed |
|
|
rs759624633 CA5883718 |
90 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs1433501817 CA379654119 |
90 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA379654097 rs1357584471 |
93 | G>E | No |
ClinGen gnomAD |
|
|
rs952257251 CA217666329 |
93 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs139582089 CA5883716 |
95 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA379654082 rs1564878799 |
96 | V>L | No |
ClinGen Ensembl |
|
|
rs749486603 CA5883715 |
101 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773633079 CA5883714 |
102 | P>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1167934257 CA379654041 |
102 | P>S | No |
ClinGen gnomAD |
|
|
rs147788904 CA5883713 |
104 | P>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA379654007 rs1482739413 |
107 | G>R | No |
ClinGen gnomAD |
|
|
CA5883711 rs781115974 |
108 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA5883709 rs183775500 |
111 | V>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA217666292 rs1055929181 |
111 | V>M | No |
ClinGen TOPMed |
|
|
CA5883708 rs779509043 |
112 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA5883707 rs757970448 |
113 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs144289891 CA5883706 |
114 | L>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA217666243 VAR_027763 rs17852369 |
116 | W>R | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs756437037 CA5883704 |
117 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA5883702 rs768023465 |
118 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA5883703 rs753037247 |
118 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA5883701 rs140328426 |
119 | P>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs573107264 CA5883700 |
122 | R>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA5883699 rs150509271 |
122 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs949259277 CA217666203 |
125 | A>E | No |
ClinGen Ensembl |
|
|
rs763181084 CA217666189 |
127 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs141810207 CA217666196 |
127 | Y>C | No |
ClinGen ESP TOPMed |
|
|
rs773508257 CA5883697 |
129 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
rs1422771238 CA379653874 |
129 | Y>H | No |
ClinGen gnomAD |
|
| TCGA novel | 132 | S>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1178638190 CA379653852 |
132 | S>P | No |
ClinGen gnomAD |
|
|
rs1352009475 CA379653176 |
134 | T>S | No |
ClinGen TOPMed |
|
|
rs374173885 CA5883678 |
135 | W>C | No |
ClinGen ESP ExAC gnomAD |
|
|
rs761918082 CA5883677 |
136 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768324196 CA5883675 |
138 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA379653148 rs768324196 |
138 | S>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA379653139 rs1301716861 |
140 | I>L | No |
ClinGen gnomAD |
|
|
rs1293101263 CA379653128 |
141 | P>L | No |
ClinGen gnomAD |
|
|
CA5883672 rs772031571 |
141 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1365331127 CA379653117 |
143 | I>F | No |
ClinGen gnomAD |
|
|
CA379653111 rs1453799245 |
144 | I>L | No |
ClinGen TOPMed gnomAD |
|
|
CA379653108 rs1387231691 |
144 | I>T | No |
ClinGen gnomAD |
|
|
CA379653112 rs1453799245 |
144 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs745336503 CA5883670 |
146 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA5883668 rs778376672 CA379653080 |
148 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5883666 rs748884292 |
150 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA5883665 rs777536488 |
152 | N>H | No |
ClinGen ExAC gnomAD |
|
|
rs747380310 CA5883663 |
153 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA5883661 rs758995339 |
156 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA379653023 rs1256378443 |
157 | T>I | No |
ClinGen gnomAD |
|
|
CA379653020 rs1232467116 |
158 | Q>* | No |
ClinGen TOPMed |
|
|
CA379653018 rs1232192253 |
158 | Q>R | No |
ClinGen gnomAD |
|
|
CA5883660 rs751015654 |
160 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA5883659 rs765199700 |
161 | E>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA217663653 rs76097537 |
162 | F>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA5883658 rs529330119 |
163 | I>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs753949467 CA5883657 |
164 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1183639004 CA379652978 |
165 | S>R | No |
ClinGen TOPMed |
|
|
CA5883655 rs760424700 |
168 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs72861055 CA5883654 |
169 | Y>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA379652947 rs1591513366 |
169 | Y>S | No |
ClinGen Ensembl |
|
|
rs188696058 CA5883653 |
170 | S>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs975543146 CA379652933 |
171 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
rs975543146 CA217663615 |
171 | V>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1327799050 CA379652923 |
173 | S>C | No |
ClinGen TOPMed |
|
|
rs1591513354 CA379652925 |
173 | S>P | No |
ClinGen Ensembl |
|
|
CA379652911 rs1591513346 |
175 | Y>S | No |
ClinGen Ensembl |
|
|
CA5883651 rs774335191 |
177 | T>P | No |
ClinGen ExAC gnomAD |
|
|
rs769515509 CA5883647 |
181 | P>H | No |
ClinGen ExAC gnomAD |
|
|
rs772670938 CA5883648 |
181 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1169287640 CA379652839 |
182 | T>I | No |
ClinGen gnomAD |
|
|
CA5883645 rs138655778 |
184 | T>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5883643 rs779489417 |
184 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA5883644 rs138655778 |
184 | T>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs779489417 CA5883642 |
184 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs370131924 CA5883640 |
185 | P>L | No |
ClinGen ESP ExAC TOPMed |
|
|
rs749336203 COSM1350607 CA5883641 |
185 | P>T | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs764175570 CA5883639 |
186 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1229136518 CA379652790 |
186 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs756309261 CA5883637 |
189 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA379652715 rs1340516763 |
190 | S>Y | No |
ClinGen gnomAD |
|
|
rs752960499 CA5883636 |
191 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA5883635 rs767300063 |
192 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA379652686 rs1213730585 |
192 | S>Y | No |
ClinGen TOPMed |
|
|
rs1199260512 CA379652677 |
193 | I>V | No |
ClinGen gnomAD |
|
|
CA217663501 rs865971373 |
194 | P>S | No |
ClinGen Ensembl |
|
|
CA5883633 rs774095385 |
195 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA5883634 rs200251557 |
195 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA379652597 rs1165579258 |
198 | K>E | No |
ClinGen gnomAD |
|
| rs1241350290 | 198 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA379652600 rs1165579258 |
198 | K>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 198 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs200474910 CA217663481 |
199 | L>M | No |
ClinGen 1000Genomes gnomAD |
|
|
rs1367939450 CA379652573 |
199 | L>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1439772218 CA379652554 |
200 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA5883631 rs766342469 |
201 | C>F | No |
ClinGen ExAC gnomAD |
|
|
rs926700705 CA217663468 |
203 | T>S | No |
ClinGen TOPMed |
|
|
CA379652494 rs1205532630 |
204 | E>Q | No |
ClinGen gnomAD |
|
|
rs912802779 CA217663454 |
205 | V>A | No |
ClinGen Ensembl |
|
|
rs376908330 CA5883630 |
207 | M>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs145069203 CA5883629 |
208 | E>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs747770568 CA5883627 |
210 | S>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747770568 CA379652380 |
210 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA217663430 rs201397416 |
210 | S>R | No |
ClinGen 1000Genomes |
|
|
CA379652352 rs1216598499 |
212 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
rs16907980 CA5883625 VAR_027764 |
214 | T>I | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs746240000 CA5883624 |
218 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA379652230 rs1435590738 |
219 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1329268618 CA379652209 |
220 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA5883623 rs373364929 |
221 | E>A | No |
ClinGen ESP ExAC gnomAD |
|
|
CA379652145 rs1275202925 |
223 | K>R | No |
ClinGen TOPMed |
|
|
rs746598821 CA217663407 |
224 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746598821 CA5883622 |
224 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5883620 rs200122188 |
226 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756257871 CA5883619 |
229 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs955203466 CA217663362 |
230 | A>G | No |
ClinGen TOPMed |
|
|
rs1029365465 CA379652061 |
231 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1029365465 CA217663358 |
231 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA379652055 rs1181695781 |
232 | G>E | No |
ClinGen gnomAD |
|
|
rs377086414 CA217663341 |
232 | G>R | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1250490225 CA379652044 |
234 | G>R | No |
ClinGen TOPMed |
|
|
rs1564877393 CA379652036 |
235 | G>R | No |
ClinGen Ensembl |
|
|
rs768456049 CA5883607 |
236 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA5883604 rs771243144 |
238 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA379651261 rs1236602144 |
240 | L>P | No |
ClinGen gnomAD |
|
|
rs1011426411 CA217662825 |
244 | A>V | No |
ClinGen TOPMed |
|
|
rs1326559895 CA379651158 |
245 | L>F | No |
ClinGen gnomAD |
|
|
rs533840110 CA5883601 |
245 | L>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA379651130 rs1268203046 |
247 | F>L | No |
ClinGen TOPMed |
|
|
CA5883598 rs140605904 |
249 | G>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1213569722 CA379651087 |
249 | G>S | No |
ClinGen TOPMed |
|
|
rs1433490448 CA379651074 |
250 | A>T | No |
ClinGen gnomAD |
|
|
rs371884025 CA217662782 |
251 | A>T | No |
ClinGen ESP TOPMed |
|
|
CA379651045 rs1366108622 |
252 | A>T | No |
ClinGen gnomAD |
|
|
CA5883597 rs755217678 |
252 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1271386448 CA379651029 |
253 | G>S | No |
ClinGen TOPMed |
|
|
CA379650998 rs1408514024 |
254 | L>R | No |
ClinGen gnomAD |
|
|
CA5883595 rs151319034 |
255 | G>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA379650992 rs1424178195 |
256 | F>I | No |
ClinGen gnomAD |
|
|
CA379650980 rs779868511 |
257 | C>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5883594 rs779868511 |
257 | C>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1006591910 CA217662726 |
258 | Y>C | No |
ClinGen Ensembl |
|
|
rs758194425 CA5883593 |
258 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA5883591 rs765270056 |
259 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1458778983 CA379650965 |
260 | K>E | No |
ClinGen TOPMed |
|
|
CA379650184 rs1390254942 |
270 | N>H | No |
ClinGen TOPMed |
|
|
CA5883564 rs767085190 |
272 | N>I | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 273 | Q>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 276 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5883562 rs773591780 CA5883563 |
277 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1330965663 CA379650114 |
277 | M>T | No |
ClinGen TOPMed |
|
|
CA5883561 rs762279093 |
278 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1282234242 CA379650109 |
278 | I>V | No |
ClinGen gnomAD |
|
|
CA5883559 rs117443264 |
279 | E>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs768769189 CA5883558 |
280 | T>N | No |
ClinGen ExAC gnomAD |
|
|
rs747073252 CA5883557 |
285 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs1351121269 CA379650064 |
285 | E>K | No |
ClinGen gnomAD |
|
|
CA5883556 rs751820193 |
286 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA5883555 rs142573261 |
286 | E>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5883554 rs746141063 |
289 | N>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 290 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1251525614 CA379650019 |
291 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
rs757182173 CA5883553 |
292 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs535742623 CA5883551 |
296 | E>Q | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1266120273 CA379649965 |
298 | K>R | No |
ClinGen gnomAD |
|
|
rs147714545 CA379649845 |
304 | P>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs147714545 CA5883548 |
304 | P>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5883547 rs767152258 |
305 | E>* | No |
ClinGen ExAC gnomAD |
|
|
rs1287416129 CA379649808 |
306 | E>G | No |
ClinGen gnomAD |
|
|
CA379649817 rs1003761554 |
306 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1003761554 CA217662213 |
306 | E>Q | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 307 | S>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs754567175 CA5883546 |
309 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA379649733 rs1457007908 |
310 | P>R | No |
ClinGen gnomAD |
|
|
rs1347591388 CA379649706 |
312 | K>Q | No |
ClinGen gnomAD |
|
|
CA5883544 rs765583615 |
312 | K>R | No |
ClinGen ExAC |
|
|
CA379649657 rs1457435979 |
314 | T>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA5883541 rs7948666 VAR_027765 |
315 | V>M | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs775409214 COSM2109480 CA5883539 |
316 | R>* | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1191090779 COSM1350603 CA379649631 |
316 | R>Q | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA379649588 rs1249979003 |
319 | E>K | No |
ClinGen gnomAD |
|
|
rs772231066 CA5883538 |
323 | V>Q | No |
ClinGen ExAC TOPMed gnomAD |
No associated diseases with Q9Y5Y7
5 regional properties for Q9Y5Y7
Functions
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| cargo receptor activity | Binding specifically to a substance (cargo) to deliver it to a transport vesicle. Cargo receptors span a membrane (either the plasma membrane or a vesicle membrane), binding simultaneously to cargo molecules and coat adaptors, to efficiently recruit soluble proteins to nascent vesicles. |
| hyaluronic acid binding | Binding to hyaluronic acid, a polymer composed of repeating dimeric units of glucuronic acid and N-acetyl glucosamine. |
| signaling receptor activity | Receiving a signal and transmitting it in the cell to initiate a change in cell activity. A signal is a physical entity or change in state that is used to transfer information in order to trigger a response. |
| transmembrane signaling receptor activity | Combining with an extracellular or intracellular signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity or state as part of signal transduction. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| anatomical structure morphogenesis | The process in which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form. |
| cell-matrix adhesion | The binding of a cell to the extracellular matrix via adhesion molecules. |
| hyaluronan catabolic process | The chemical reactions and pathways resulting in the breakdown of hyaluronan, the naturally occurring anionic form of hyaluronic acid, any member of a group of glycosaminoglycans, the repeat units of which consist of beta-1,4 linked D-glucuronyl-beta-(1,3)-N-acetyl-D-glucosamine. |
| positive regulation of cellular extravasation | Any process that activates or increases the frequency, rate, or extent of cellular extravasation. |
| receptor-mediated endocytosis | An endocytosis process in which cell surface receptors ensure specificity of transport. A specific receptor on the cell surface binds tightly to the extracellular macromolecule (the ligand) that it recognizes; the plasma-membrane region containing the receptor-ligand complex then undergoes endocytosis, forming a transport vesicle containing the receptor-ligand complex and excluding most other plasma-membrane proteins. Receptor-mediated endocytosis generally occurs via clathrin-coated pits and vesicles. |
| response to wounding | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q6UC88 | LYVE1 | Lymphatic vessel endothelial hyaluronic acid receptor 1 | Bos taurus (Bovine) | PR |
| P98066 | TNFAIP6 | Tumor necrosis factor-inducible gene 6 protein | Homo sapiens (Human) | PR |
| Q8BHC0 | Lyve1 | Lymphatic vessel endothelial hyaluronic acid receptor 1 | Mus musculus (Mouse) | PR |
| O08859 | Tnfaip6 | Tumor necrosis factor-inducible gene 6 protein | Mus musculus (Mouse) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MARCFSLVLL | LTSIWTTRLL | VQGSLRAEEL | SIQVSCRIMG | ITLVSKKANQ | QLNFTEAKEA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CRLLGLSLAG | KDQVETALKA | SFETCSYGWV | GDGFVVISRI | SPNPKCGKNG | VGVLIWKVPV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SRQFAAYCYN | SSDTWTNSCI | PEIITTKDPI | FNTQTATQTT | EFIVSDSTYS | VASPYSTIPA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PTTTPPAPAS | TSIPRRKKLI | CVTEVFMETS | TMSTETEPFV | ENKAAFKNEA | AGFGGVPTAL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LVLALLFFGA | AAGLGFCYVK | RYVKAFPFTN | KNQQKEMIET | KVVKEEKAND | SNPNEESKKT |
| 310 | 320 | ||||
| DKNPEESKSP | SKTTVRCLEA | EV |