Q9Y5S2
Gene name |
CDC42BPB (KIAA1124) |
Protein name |
Serine/threonine-protein kinase MRCK beta |
Names |
EC 2.7.11.1 , CDC42-binding protein kinase beta , CDC42BP-beta , DMPK-like beta , Myotonic dystrophy kinase-related CDC42-binding kinase beta , MRCK beta , Myotonic dystrophy protein kinase-like beta |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9578 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
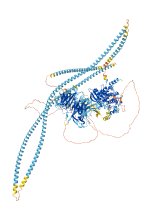
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
76-342 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
217-241 (Activation loop from InterPro)
Target domain |
3-411 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1543 variants for Q9Y5S2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001007682 rs1595208914 |
13 | L>missing | CDC42BPB-related neurodevelopmental syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1384767070 RCV001839070 |
44 | S>L | CDC42BPB-related Neurodevelopmental disorder [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
RCV001007673 rs1198710710 VAR_087124 |
142 | A>T | CDC42BPB-related neurodevelopmental syndrome CHOCNS; uncertain significance [ClinVar, UniProt] | Yes |
ClinVar TOPMed dbSNP gnomAD UniProt |
|
RCV002249553 rs1595127294 RCV001007674 VAR_087125 RCV003127490 |
175 | D>Y | CDC42BPB-related neurodevelopmental syndrome Chilton-Okur-Chung neurodevelopmental syndrome Autism spectrum disorder CHOCNS; uncertain significance [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
VAR_087126 RCV002251523 RCV001007675 rs1016320330 |
293 | I>M | CDC42BPB-related neurodevelopmental syndrome Chilton-Okur-Chung neurodevelopmental syndrome CHOCNS; uncertain significance [ClinVar, UniProt] | Yes |
ClinVar dbSNP gnomAD UniProt |
|
RCV001266866 rs1389878014 |
512 | E>Q | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
RCV001007683 rs1595482918 |
544 | E>missing | CDC42BPB-related neurodevelopmental syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001007684 rs1595476797 RCV002249556 |
764 | R>* | CDC42BPB-related neurodevelopmental syndrome Chilton-Okur-Chung neurodevelopmental syndrome [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
| VAR_087127 | 764 | R>del | CHOCNS [UniProt] | Yes | UniProt |
|
COSM348876 RCV002051901 VAR_087128 RCV002249554 RCV001007676 RCV001585796 rs1595472764 |
867 | R>C | lung CDC42BPB-related neurodevelopmental syndrome CDC42BPB-related disorder Chilton-Okur-Chung neurodevelopmental syndrome CHOCNS [Cosmic, ClinVar, UniProt] | Yes |
cosmic curated ClinVar Ensembl dbSNP UniProt |
|
rs1595472756 RCV001007677 VAR_087129 |
871 | L>P | CDC42BPB-related neurodevelopmental syndrome CHOCNS; uncertain significance [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
RCV001007679 rs1595472739 VAR_087130 |
876 | R>P | CDC42BPB-related neurodevelopmental syndrome CHOCNS [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
RCV001007678 RCV002249555 COSM21311 rs1595472741 |
876 | R>W | CDC42BPB-related neurodevelopmental syndrome large_intestine Chilton-Okur-Chung neurodevelopmental syndrome [ClinVar, Cosmic] | Yes |
cosmic curated ClinVar Ensembl dbSNP |
|
RCV001650496 rs1346688931 |
1205 | W>* | Neurodevelopmental disorder [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
rs1595450393 VAR_087131 RCV001007680 |
1299 | R>Q | CDC42BPB-related neurodevelopmental syndrome CHOCNS; uncertain significance [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
rs1595450125 VAR_087132 RCV001007681 |
1350 | R>P | CDC42BPB-related neurodevelopmental syndrome CHOCNS; uncertain significance [ClinVar, UniProt] | Yes |
ClinVar UniProt Ensembl dbSNP |
| rs1221606316 | 2 | S>L | No |
TOPMed gnomAD |
|
| rs1889033453 | 3 | A>S | No | Ensembl | |
| rs1889033282 | 3 | A>V | No | gnomAD | |
| TCGA novel | 4 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1331211983 | 4 | K>R | No | gnomAD | |
| rs1288920926 | 6 | R>W | No |
TOPMed gnomAD |
|
| rs749933054 | 8 | K>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1335725420 | 8 | K>R | No |
TOPMed gnomAD |
|
| rs1407264836 | 12 | Q>R | No |
TOPMed gnomAD |
|
| rs1889031169 | 14 | L>F | No |
TOPMed gnomAD |
|
| rs757691424 | 16 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1889030348 | 16 | D>G | No | Ensembl | |
| rs1160560731 | 16 | D>H | No | gnomAD | |
| rs752178375 | 18 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1241074729 | 19 | W>G | No | TOPMed | |
| rs201340035 | 20 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1889029511 | 21 | N>H | No | TOPMed | |
| rs1242282334 | 21 | N>S | No |
TOPMed gnomAD |
|
| rs1232684545 | 23 | S>N | No | Ensembl | |
| rs1466090550 | 23 | S>R | No | gnomAD | |
| rs1889028716 | 24 | A>V | No | TOPMed | |
| rs370755673 | 29 | T>A | No |
ESP gnomAD |
|
| rs763422325 | 33 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs752916061 | 34 | L>F | No |
ExAC gnomAD |
|
| rs1450828740 | 35 | V>A | No |
TOPMed gnomAD |
|
| rs759838637 | 39 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs992747877 | 42 | S>N | No | TOPMed | |
| rs1164936094 | 42 | S>R | No | TOPMed | |
| rs1425392018 | 43 | H>R | No |
TOPMed gnomAD |
|
| rs1889025433 | 43 | H>Y | No | Ensembl | |
| rs1419664091 | 45 | A>V | No |
TOPMed gnomAD |
|
|
COSM3401124 rs1248462707 |
47 | R>C | central_nervous_system [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1248462707 | 47 | R>G | No |
TOPMed gnomAD |
|
| rs1889023648 | 49 | D>V | No | TOPMed | |
| rs1392880438 | 49 | D>Y | No |
TOPMed gnomAD |
|
| rs768383971 | 50 | K>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs774279882 | 50 | K>Q | No |
ExAC TOPMed |
|
| rs1291386827 | 51 | Y>H | No | gnomAD | |
| rs959796421 | 52 | V>A | No |
TOPMed gnomAD |
|
| rs749105563 | 53 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs769301150 | 57 | E>K | No |
ExAC gnomAD |
|
| rs769301150 | 57 | E>Q | No |
ExAC gnomAD |
|
| rs1889021474 | 58 | W>L | No | TOPMed | |
| COSM4049247 | 59 | A>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs995127355 | 59 | A>T | No |
TOPMed gnomAD |
|
| rs775126793 | 60 | K>E | No |
ExAC TOPMed gnomAD |
|
| COSM953610 | 60 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1886193515 | 61 | P>S | No | Ensembl | |
| rs769695479 | 63 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs780909971 | 64 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1886192116 | 66 | V>M | No | Ensembl | |
| rs552932246 | 67 | K>R | No | 1000Genomes | |
| rs1480673716 | 69 | M>I | No |
TOPMed gnomAD |
|
| rs2139642174 | 70 | Q>R | No | Ensembl | |
| rs1886191319 | 71 | L>F | No | Ensembl | |
| rs1886191153 | 72 | H>R | No | TOPMed | |
| rs777461445 | 73 | R>Q | No |
ExAC gnomAD |
|
| COSM271015 | 75 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3493952 | 75 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2139642122 | 77 | E>D | No | Ensembl | |
| rs1202032028 | 77 | E>K | No | gnomAD | |
| rs2139642101 | 83 | G>R | No | Ensembl | |
| rs1886190549 | 88 | G>D | No | Ensembl | |
| TCGA novel | 89 | E>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3814284 | 91 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764882406 | 94 | K>R | No |
ExAC gnomAD |
|
| rs1319097165 | 98 | T>A | No | gnomAD | |
| rs1885974911 | 99 | E>K | No | TOPMed | |
| rs2139628971 | 100 | R>* | No | Ensembl | |
| rs776104283 | 100 | R>L | No |
ExAC gnomAD |
|
|
COSM4049246 rs776104283 |
100 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| COSM4049245 | 101 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772832755 | 107 | L>F | No |
ExAC gnomAD |
|
| rs1885973429 | 108 | N>K | No | TOPMed | |
| rs1020400206 | 108 | N>S | No | TOPMed | |
|
RCV001771412 rs1171933717 |
111 | E>A | No |
ClinVar Ensembl dbSNP |
|
| rs771777803 | 111 | E>D | No |
ExAC gnomAD |
|
| rs1171933717 | 111 | E>G | No | Ensembl | |
| rs748539078 | 117 | E>D | No |
ExAC gnomAD |
|
| rs1185305624 | 119 | A>S | No |
TOPMed gnomAD |
|
| rs1185305624 | 119 | A>T | No |
TOPMed gnomAD |
|
|
RCV001380967 rs2139609624 |
122 | R>* | No |
ClinVar Ensembl dbSNP |
|
|
COSM697009 rs1895116700 |
122 | R>Q | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs1357631483 | 123 | E>Q | No | gnomAD | |
|
rs932105669 COSM3493951 |
125 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs781414650 | 126 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs371426927 | 130 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1225331202 | 131 | G>R | No | gnomAD | |
| rs1225331202 | 131 | G>S | No | gnomAD | |
| rs80317877 | 132 | D>A | No | Ensembl | |
| rs777760721 | 132 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs978392521 | 133 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1386982720 | 134 | Q>R | No | TOPMed | |
| rs754926991 | 137 | T>I | No |
ExAC gnomAD |
|
| rs760562644 | 138 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs766213299 | 138 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs760562644 | 138 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1895113283 | 140 | H>R | No | TOPMed | |
| rs1481742916 | 140 | H>Y | No | gnomAD | |
| rs1489882007 | 142 | A>G | No | gnomAD | |
| rs1489882007 | 142 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1196626669 | 144 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1196626669 | 144 | Q>K | No |
TOPMed gnomAD |
|
| rs768111592 | 146 | E>K | No |
ExAC gnomAD |
|
| rs1895112018 | 147 | N>T | No | Ensembl | |
| rs1895111872 | 148 | H>D | No | TOPMed | |
| rs1895111739 | 148 | H>Q | No | Ensembl | |
| rs1895111596 | 149 | L>P | No | gnomAD | |
| rs1894893081 | 152 | V>A | No | TOPMed | |
| rs773322928 | 152 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs773322928 | 152 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1894892811 | 155 | Y>F | No | TOPMed | |
| rs748273726 | 158 | G>D | No |
ExAC gnomAD |
|
| rs1280833656 | 158 | G>S | No | TOPMed | |
| rs1246324934 | 163 | T>I | No | gnomAD | |
| rs1246324934 | 163 | T>N | No | gnomAD | |
| TCGA novel | 165 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1894891478 | 167 | K>R | No | gnomAD | |
| COSM3493949 | 169 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs569036151 | 169 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs2139593221 | 173 | P>L | No | 1000Genomes | |
| rs200047302 | 173 | P>S | No | TOPMed | |
| rs1041133482 | 176 | M>T | No | TOPMed | |
| rs1161181768 | 179 | F>* | No | gnomAD | |
| rs1595127260 | 179 | F>V | No | Ensembl | |
| TCGA novel | 183 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760028278 | 187 | A>V | No |
ExAC gnomAD |
|
| COSM432699 | 193 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777005233 | 193 | Q>R | No |
ExAC gnomAD |
|
| rs147856179 | 197 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs147856179 | 197 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs957253615 | 202 | K>R | No | TOPMed | |
| TCGA novel | 203 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs62006862 | 203 | P>L | No | Ensembl | |
| rs62006861 | 204 | D>Y | No | Ensembl | |
| rs1476567278 | 205 | N>S | No |
TOPMed gnomAD |
|
| rs773431264 | 210 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs1273277947 | 210 | V>M | No |
TOPMed gnomAD |
|
| TCGA novel | 210 | V>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1894254247 | 211 | N>K | No | Ensembl | |
| rs762122928 | 215 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs774720497 | 215 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| TCGA novel | 215 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1357642794 | 217 | A>G | No | TOPMed | |
| rs112134218 | 219 | F>S | No | Ensembl | |
| rs1347313535 | 221 | S>L | No | gnomAD | |
| rs1313733252 | 225 | M>I | No | TOPMed | |
| COSM4049243 | 227 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1894252552 | 228 | D>G | No | gnomAD | |
| rs1468791260 | 228 | D>N | No | gnomAD | |
| rs1468791260 | 228 | D>Y | No | gnomAD | |
| rs1377307978 | 229 | G>A | No |
TOPMed gnomAD |
|
| rs1894112812 | 232 | Q>H | No |
TOPMed gnomAD |
|
| rs1894112922 | 232 | Q>R | No | TOPMed | |
| rs1184748794 | 235 | V>L | No | gnomAD | |
| rs1184748794 | 235 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1217960770 | 243 | I>L | No |
TOPMed gnomAD |
|
| rs1287926437 | 243 | I>M | No | TOPMed | |
| rs1217960770 | 243 | I>V | No |
TOPMed gnomAD |
|
| rs967740106 | 250 | A>V | No |
TOPMed gnomAD |
|
| rs779426560 | 251 | M>T | No |
ExAC gnomAD |
|
| rs1233852375 | 254 | G>D | No |
TOPMed gnomAD |
|
| rs750838520 | 254 | G>S | No |
ExAC gnomAD |
|
| rs1430759266 | 255 | M>L | No | gnomAD | |
| rs1471335993 | 257 | K>R | No |
1000Genomes TOPMed gnomAD |
|
| TCGA novel | 258 | Y>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 259 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757877062 | 261 | E>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 264 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1164545056 | 269 | V>I | No | gnomAD | |
| rs1423604188 | 271 | M>V | No |
TOPMed gnomAD |
|
| rs372288260 | 274 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1162161745 | 275 | L>I | No | Ensembl | |
| rs531412667 | 276 | Y>C | No | 1000Genomes | |
| rs1405592074 | 278 | E>K | No | Ensembl | |
| rs1486224985 | 279 | T>M | No |
TOPMed gnomAD |
|
| rs2139527081 | 283 | A>V | No | Ensembl | |
| rs1894107413 | 285 | S>T | No |
TOPMed gnomAD |
|
| rs767414589 | 287 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs767414589 | 287 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs202202441 | 289 | T>S | No |
1000Genomes ExAC gnomAD |
|
| rs768567118 | 290 | Y>F | No |
ExAC gnomAD |
|
| rs775222428 | 291 | G>A | No |
ExAC gnomAD |
|
| rs549614523 | 294 | M>V | No | Ensembl | |
| rs769139672 | 296 | H>Q | No |
ExAC gnomAD |
|
| rs1282644689 | 296 | H>R | No | TOPMed | |
| rs956631106 | 297 | E>G | No |
TOPMed gnomAD |
|
| rs1893972655 | 298 | E>* | No | Ensembl | |
| rs941463713 | 299 | R>Q | No | TOPMed | |
| rs1424438542 | 301 | Q>H | No |
TOPMed gnomAD |
|
| rs888491399 | 302 | F>L | No | TOPMed | |
| rs762982831 | 305 | H>P | No | Ensembl | |
| rs138477914 | 307 | T>M | No |
ESP gnomAD |
|
| rs776272982 | 309 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1323587983 | 311 | E>K | No |
TOPMed gnomAD |
|
| rs1893971392 | 311 | E>V | No | Ensembl | |
|
rs1893971251 TCGA novel |
313 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1893970971 | 314 | K>R | No | Ensembl | |
| rs547502635 | 321 | I>L | No |
1000Genomes ExAC gnomAD |
|
| rs547502635 | 321 | I>V | No |
1000Genomes ExAC gnomAD |
|
| rs775066402 | 323 | S>G | No |
ExAC gnomAD |
|
| rs755180952 | 323 | S>N | No |
ExAC gnomAD |
|
| rs1893969500 | 324 | R>I | No | Ensembl | |
| rs1893969237 | 325 | E>K | No | Ensembl | |
| rs754126714 | 326 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs771743362 | 326 | R>H | No |
ExAC gnomAD |
|
| rs771743362 | 326 | R>L | No |
ExAC gnomAD |
|
| rs1277252912 | 327 | R>Q | No |
TOPMed gnomAD |
|
| rs1893968413 | 330 | Q>R | No | Ensembl | |
| TCGA novel | 331 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1212092841 | 332 | G>A | No |
TOPMed gnomAD |
|
| rs34076876 | 334 | E>K | No |
ExAC gnomAD |
|
| rs34076876 | 334 | E>Q | No |
ExAC gnomAD |
|
| rs1249865429 | 335 | D>A | No | gnomAD | |
| TCGA novel | 338 | K>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 339 | H>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV001542456 rs2139515520 |
339 | H>R | No |
ClinVar Ensembl dbSNP |
|
|
rs368416667 COSM4049241 |
340 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1260922744 | 341 | F>L | No | gnomAD | |
| COSM1368393 | 342 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM273772 | 343 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 344 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1424486232 | 349 | N>H | No |
TOPMed gnomAD |
|
| TCGA novel | 349 | N>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1248642512 | 349 | N>S | No |
TOPMed gnomAD |
|
|
rs529056427 TCGA novel |
351 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA 1000Genomes |
| rs529056427 | 351 | R>G | No | 1000Genomes | |
|
COSM274460 rs772636121 |
351 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1176003660 | 352 | N>D | No |
TOPMed gnomAD |
|
| rs1893964881 | 354 | E>G | No | gnomAD | |
| rs772612460 | 354 | E>Q | No |
ExAC gnomAD |
|
|
rs1893964617 RCV001754909 |
357 | Y>* | No |
ClinVar TOPMed dbSNP |
|
| rs1893964487 | 358 | I>T | No | TOPMed | |
| COSM953606 | 363 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1893964308 | 363 | S>N | No |
TOPMed gnomAD |
|
| rs1893964308 | 363 | S>T | No |
TOPMed gnomAD |
|
| rs1893964192 | 364 | P>A | No | TOPMed | |
| COSM4049240 | 364 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3401123 | 366 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1893963310 | 371 | D>N | No | TOPMed | |
| rs1445044341 | 372 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1595490349 | 373 | D>G | No | Ensembl | |
|
COSM3983113 RCV001785216 rs1185383388 |
375 | D>N | ovary [Cosmic] | No |
cosmic curated ClinVar TOPMed dbSNP gnomAD |
| rs770798082 | 376 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs758348207 | 377 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1487065711 | 380 | T>M | No | gnomAD | |
| rs1893843484 | 382 | I>V | No |
TOPMed gnomAD |
|
|
RCV002267310 rs1437531053 |
388 | H>Q | No |
ClinVar TOPMed dbSNP gnomAD |
|
| TCGA novel | 388 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759102366 | 389 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs748069736 | 391 | F>L | No |
ExAC gnomAD |
|
| rs1400880323 | 394 | L>F | No | gnomAD | |
| rs1169912257 | 395 | H>R | No | gnomAD | |
| rs1432013256 | 395 | H>Y | No | TOPMed | |
| rs1371299187 | 399 | I>V | No |
TOPMed gnomAD |
|
| rs1893841522 | 400 | G>D | No | TOPMed | |
| rs1893841405 | 402 | T>S | No | TOPMed | |
| rs1893841162 | 405 | T>A | No | Ensembl | |
| rs1477528246 | 405 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs763381498 | 408 | C>Y | No |
ExAC gnomAD |
|
| rs1893726665 | 410 | S>C | No | Ensembl | |
|
COSM107988 rs143236725 |
412 | R>* | Variant assessed as Somatic; HIGH impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs775986875 | 412 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1193231685 | 413 | G>S | No |
TOPMed gnomAD |
|
| rs1893725711 | 414 | S>F | No | Ensembl | |
| rs1389670141 | 415 | L>P | No | TOPMed | |
| rs777092253 | 418 | I>L | No |
ExAC gnomAD |
|
| rs1319868403 | 419 | M>T | No | gnomAD | |
| rs1005957568 | 419 | M>V | No | TOPMed | |
| rs771034761 | 420 | Q>R | No |
ExAC gnomAD |
|
| rs747311960 | 421 | S>P | No |
ExAC gnomAD |
|
| rs1416537335 | 422 | N>K | No | gnomAD | |
| rs773422657 | 422 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1893724651 | 422 | N>Y | No | Ensembl | |
| rs1240863944 | 424 | L>S | No |
TOPMed gnomAD |
|
| rs1182992498 | 425 | T>A | No | gnomAD | |
| rs1893723606 | 427 | D>E | No | Ensembl | |
| rs749179808 | 427 | D>G | No | ExAC | |
| TCGA novel | 427 | D>M | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM432698 | 428 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780164492 | 429 | D>N | No |
ExAC gnomAD |
|
| TCGA novel | 429 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1357473697 | 431 | Q>R | No | TOPMed | |
| rs377230665 | 432 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4049239 rs769595230 |
432 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs781091895 | 433 | D>V | No |
ExAC gnomAD |
|
| rs532655051 | 436 | H>L | No | gnomAD | |
| rs756849826 | 436 | H>Q | No |
ExAC gnomAD |
|
| rs1227731562 | 437 | S>N | No | gnomAD | |
| rs202198663 | 440 | M>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1566874246 | 442 | A>P | No | Ensembl | |
| rs1566874246 | 442 | A>S | No | Ensembl | |
|
TCGA novel rs1893721853 |
442 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1489501101 | 443 | Y>F | No | Ensembl | |
| rs1295995685 | 444 | E>D | No | gnomAD | |
| rs753158900 | 444 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1250404274 | 448 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
|
COSM3493948 rs765630243 |
448 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1893720523 | 449 | R>K | No | TOPMed | |
| rs1420073737 | 453 | E>D | No | TOPMed | |
| rs750461988 | 459 | R>W | No | gnomAD | |
| rs1003129061 | 461 | L>P | No |
TOPMed gnomAD |
|
| COSM3671864 | 462 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1128330 | 463 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3493947 | 464 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1893712466 | 464 | S>P | No | TOPMed | |
| rs1234783943 | 465 | T>I | No |
TOPMed gnomAD |
|
| rs1234783943 | 465 | T>S | No |
TOPMed gnomAD |
|
| rs1300188295 | 467 | T>I | No | gnomAD | |
| rs759330821 | 468 | V>L | No |
ExAC gnomAD |
|
| rs759330821 | 468 | V>M | No |
ExAC gnomAD |
|
| rs1448257033 | 471 | L>R | No | gnomAD | |
| rs1893710704 | 473 | G>D | No | TOPMed | |
| rs1893710830 | 473 | G>R | No | gnomAD | |
| rs1893710830 | 473 | G>S | No | gnomAD | |
| rs371772936 | 476 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs367865440 | 476 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs367865440 | 476 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs371772936 | 476 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1015324528 | 477 | A>T | No |
TOPMed gnomAD |
|
| rs1893709744 | 479 | S>G | No | TOPMed | |
| rs754531547 | 480 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1212883584 | 482 | N>D | No | TOPMed | |
|
rs749711259 COSM953605 |
483 | R>Q | lung Variant assessed as Somatic; MODERATE impact. endometrium [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1457212134 | 486 | E>D | No | gnomAD | |
| COSM697010 | 486 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1236298714 | 487 | I>M | No | gnomAD | |
| rs1201621767 | 488 | K>Q | No | gnomAD | |
| rs185643206 | 488 | K>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM3814283 | 489 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1255018308 | 490 | L>P | No | gnomAD | |
| rs756727099 | 491 | N>D | No |
ExAC gnomAD |
|
| rs530300961 | 491 | N>S | No |
TOPMed gnomAD |
|
| rs530300961 | 491 | N>T | No |
TOPMed gnomAD |
|
| rs750798248 | 493 | E>D | No |
ExAC gnomAD |
|
| rs377142500 | 494 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1893707085 | 494 | I>T | No | Ensembl | |
| rs768057349 | 494 | I>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 495 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751814078 | 495 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs140317521 | 496 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs372371951 | 496 | R>H | No |
ESP TOPMed gnomAD |
|
| rs112025674 | 497 | L>F | No | TOPMed | |
| COSM953604 | 498 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs773352644 | 498 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs775468568 | 499 | N>K | No |
ExAC gnomAD |
|
|
rs1158962067 VAR_040834 COSM619 |
500 | K>E | breast a breast infiltrating ductal carcinoma sample; somatic mutation [Cosmic, UniProt] | No |
cosmic curated UniProt dbSNP gnomAD |
| rs766277875 | 501 | I>M | No |
ExAC gnomAD |
|
| rs1231370027 | 501 | I>V | No |
TOPMed gnomAD |
|
| rs1566873808 | 502 | A>T | No | TOPMed | |
| rs1261899684 | 502 | A>V | No |
TOPMed gnomAD |
|
| rs1212006380 | 503 | D>V | No |
TOPMed gnomAD |
|
| rs1893623620 | 504 | S>L | No |
TOPMed gnomAD |
|
| rs773145635 | 506 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1893623174 | 507 | L>F | No | TOPMed | |
| rs1893623023 | 507 | L>P | No | Ensembl | |
| rs774054692 | 508 | E>D | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 508 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768333860 | 509 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs768333860 | 509 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs762573744 | 509 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1387162153 | 510 | Q>P | No | gnomAD | |
| COSM3793489 | 512 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs769436597 | 512 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs746321865 | 514 | T>A | No |
ExAC gnomAD |
|
| rs143474150 | 514 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1595483123 | 515 | V>G | No | Ensembl | |
| rs1476597501 | 516 | A>T | No |
TOPMed gnomAD |
|
|
rs149124468 COSM2151603 |
516 | A>V | central_nervous_system [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1893620425 | 517 | L>P | No | gnomAD | |
| rs200810382 | 518 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201875902 | 518 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201875902 | 518 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1056905207 | 521 | R>C | No |
TOPMed gnomAD |
|
| rs1056905207 | 521 | R>G | No |
TOPMed gnomAD |
|
|
rs367791222 COSM4049238 |
521 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs367791222 | 521 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs917093626 | 523 | D>A | No |
TOPMed gnomAD |
|
| rs917093626 | 523 | D>V | No |
TOPMed gnomAD |
|
| rs767412929 | 524 | S>P | No |
ExAC gnomAD |
|
| rs761707317 | 525 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1323795391 | 526 | Q>R | No |
TOPMed gnomAD |
|
| rs776779318 | 527 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs776779318 | 527 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1448994494 | 527 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs200053443 | 529 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200053443 | 529 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1163047 rs374111285 |
529 | R>W | pancreas [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs147371660 | 530 | G>R | No |
ESP gnomAD |
|
| rs1192156263 | 530 | G>V | No | gnomAD | |
| rs1893616531 | 531 | L>Q | No | Ensembl | |
| rs1289000508 | 533 | K>R | No |
TOPMed gnomAD |
|
| rs776328032 | 535 | H>Q | No |
ExAC gnomAD |
|
| rs1566872144 | 535 | H>Y | No | gnomAD | |
| rs771452351 | 536 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs144313805 | 536 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs144313805 | 536 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs140070781 | 537 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs964090318 | 538 | V>A | No | gnomAD | |
| rs1377486618 | 538 | V>F | No |
TOPMed gnomAD |
|
| rs1377486618 | 538 | V>I | No |
TOPMed gnomAD |
|
| rs755233914 | 539 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs753894689 | 539 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs753894689 | 539 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs755233914 | 539 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs757217270 | 542 | K>E | No |
ExAC gnomAD |
|
| rs1413616750 | 542 | K>R | No | Ensembl | |
| rs751458409 | 544 | E>* | No |
ExAC TOPMed gnomAD |
|
|
rs763896010 RCV001774676 |
544 | E>G | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs751458409 | 544 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs763896010 | 544 | E>V | No |
ExAC gnomAD |
|
| rs1468655007 | 546 | H>Q | No | gnomAD | |
| rs752576729 | 546 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1893612312 | 547 | K>E | No | Ensembl | |
| rs1239356043 | 547 | K>M | No | gnomAD | |
| rs2139475497 | 550 | V>A | No | Ensembl | |
| rs1255101835 | 550 | V>I | No | TOPMed | |
| rs1893502669 | 554 | E>Q | No | Ensembl | |
|
rs36001612 VAR_040835 |
555 | R>Q | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs931183204 | 555 | R>W | No |
TOPMed gnomAD |
|
| TCGA novel | 558 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374954708 | 561 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs749456768 | 561 | K>R | No |
ExAC gnomAD |
|
| rs2139475387 | 562 | E>* | No | Ensembl | |
| rs550942756 | 562 | E>D | No |
1000Genomes ExAC gnomAD |
|
| rs931987662 | 564 | K>R | No |
TOPMed gnomAD |
|
| TCGA novel | 565 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200151871 | 566 | A>V | No | Ensembl | |
| rs1893500560 | 567 | H>R | No | Ensembl | |
| rs1288598764 | 567 | H>Y | No | gnomAD | |
| TCGA novel | 570 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs148385475 | 570 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1374958981 | 572 | L>M | No | gnomAD | |
| rs1374958981 | 572 | L>V | No | gnomAD | |
| rs537535669 | 573 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs1166159751 | 573 | A>V | No | gnomAD | |
| rs1370551078 | 575 | Q>R | No | TOPMed | |
| rs1415841205 | 577 | F>I | No |
TOPMed gnomAD |
|
| rs1415841205 | 577 | F>L | No |
TOPMed gnomAD |
|
| TCGA novel | 578 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1261329799 | 580 | L>V | No |
TOPMed gnomAD |
|
| rs149554853 | 581 | N>D | No |
ESP TOPMed |
|
| TCGA novel | 581 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778791236 | 581 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs754841354 | 582 | E>G | No |
ExAC gnomAD |
|
| rs1489366169 | 582 | E>K | No | gnomAD | |
| rs141131354 | 583 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs765917838 | 583 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1218724775 | 585 | A>T | No | gnomAD | |
| rs1893497396 | 586 | E>D | No | gnomAD | |
| rs2139474990 | 587 | L>F | No | Ensembl | |
| rs372968163 | 588 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs551755307 | 588 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs1893496826 | 589 | A>D | No | TOPMed | |
| rs767081811 | 590 | Q>K | No |
ExAC gnomAD |
|
| rs1370240777 | 590 | Q>L | No |
TOPMed gnomAD |
|
| COSM3987521 | 591 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1893496402 | 591 | K>Q | No | gnomAD | |
| rs2139474869 | 595 | S>F | No | Ensembl | |
| rs775968869 | 596 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs763342575 | 596 | R>W | No |
ExAC TOPMed |
|
| rs1893495301 | 598 | L>P | No | TOPMed | |
|
rs369423884 COSM174688 |
599 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs759867170 | 600 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1011557884 | 600 | D>G | No | gnomAD | |
| rs1011557884 | 600 | D>V | No | gnomAD | |
| rs894014625 | 601 | K>E | No |
TOPMed gnomAD |
|
| rs776505275 | 602 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1192779388 | 603 | E>K | No | gnomAD | |
| rs1031580687 | 605 | M>T | No | Ensembl | |
| rs372499348 | 605 | M>V | No |
ESP TOPMed gnomAD |
|
| rs748047064 | 607 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs748047064 | 607 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs868554043 | 608 | A>D | No | Ensembl | |
| rs779024334 | 608 | A>T | No |
ExAC gnomAD |
|
| rs868554043 | 608 | A>V | No | Ensembl | |
| rs145670721 | 609 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs145670721 | 609 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1893492563 | 610 | Q>R | No |
TOPMed gnomAD |
|
| rs1893491970 | 614 | A>D | No |
TOPMed gnomAD |
|
|
rs1893492115 COSM6025538 |
614 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic gnomAD |
| rs1380140937 | 615 | M>V | No |
TOPMed gnomAD |
|
| rs1369378750 | 616 | R>Q | No |
TOPMed gnomAD |
|
| rs780626139 | 616 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| COSM1368392 | 617 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1380110869 | 617 | Q>L | No | Ensembl | |
| rs140632643 | 620 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs761404232 | 620 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs947730985 | 621 | R>K | No | gnomAD | |
| rs1359980854 | 626 | R>G | No | gnomAD | |
| rs763569929 | 627 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1893416610 | 630 | E>K | No | TOPMed | |
| rs1362004091 | 631 | A>S | No | gnomAD | |
| rs1362004091 | 631 | A>T | No | gnomAD | |
| rs1188959808 | 631 | A>V | No | gnomAD | |
| rs751052410 | 633 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs772855288 | 634 | D>G | No | gnomAD | |
| rs372304425 | 634 | D>H | No |
ESP TOPMed |
|
| rs1893415071 | 635 | D>G | No | gnomAD | |
| rs777327728 | 635 | D>H | No |
ExAC gnomAD |
|
| rs777327728 | 635 | D>N | No |
ExAC gnomAD |
|
| rs368705809 | 637 | V>I | No |
ESP ExAC gnomAD |
|
| rs1893414721 | 638 | A>G | No | TOPMed | |
| rs373805829 | 642 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1249937943 | 644 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| COSM1368391 | 644 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1893413763 | 645 | K>R | No | TOPMed | |
| rs1247042382 | 646 | L>F | No | gnomAD | |
| rs1247042382 | 646 | L>V | No | gnomAD | |
|
rs754293915 COSM953603 |
647 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs766686530 | 649 | H>R | No |
ExAC gnomAD |
|
| rs1437457747 | 650 | S>G | No |
TOPMed gnomAD |
|
|
COSM953602 rs761037229 |
650 | S>N | endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1893412768 | 651 | E>D | No | TOPMed | |
| rs138666514 | 651 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs138666514 | 651 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1318657834 | 653 | F>L | No |
TOPMed gnomAD |
|
| rs1365551712 | 653 | F>L | No | TOPMed | |
| rs1381459338 | 656 | Q>R | No | gnomAD | |
| rs1893411666 | 659 | S>G | No | Ensembl | |
| rs559189691 | 659 | S>N | No |
1000Genomes ExAC gnomAD |
|
| rs769438435 | 660 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1167097758 | 663 | A>T | No |
TOPMed gnomAD |
|
| rs1162177742 | 663 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs745647838 | 665 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs540629746 | 666 | V>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs540629746 | 666 | V>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs560275092 | 666 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs374881719 | 667 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1282604230 | 667 | K>R | No | gnomAD | |
| rs778344538 | 668 | Q>E | No |
ExAC gnomAD |
|
| rs1276334854 | 668 | Q>H | No | gnomAD | |
| rs947629970 | 669 | G>R | No |
TOPMed gnomAD |
|
| rs144812182 | 671 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
VAR_040836 rs55948035 RCV002248322 |
671 | R>Q | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs144812182 | 671 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1893334283 COSM337379 |
672 | G>A | lung [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1893334412 | 672 | G>R | No | TOPMed | |
| rs750636528 | 673 | A>V | No |
ExAC gnomAD |
|
| rs1893333779 | 674 | G>V | No | Ensembl | |
| rs1407786828 | 679 | H>Q | No | gnomAD | |
| rs1595477267 | 680 | Q>K | No | Ensembl | |
| rs751650285 | 681 | Q>E | No |
ExAC gnomAD |
|
| TCGA novel | 682 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs542009619 | 682 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs753792492 | 687 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1893332407 | 688 | S>C | No | Ensembl | |
|
rs1893332407 RCV001542378 |
688 | S>F | No |
ClinVar Ensembl dbSNP |
|
| rs1471198756 | 689 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 691 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772927977 | 693 | K>N | No |
ExAC gnomAD |
|
| TCGA novel | 696 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1893331509 | 696 | F>S | No | Ensembl | |
| rs771742248 | 697 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs771742248 | 697 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs1566866914 | 699 | E>A | No | Ensembl | |
| rs761439565 | 699 | E>D | No |
ExAC gnomAD |
|
| COSM432697 | 699 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs912861997 | 700 | E>K | No | TOPMed | |
| rs1211297916 | 701 | L>F | No | gnomAD | |
| rs1377141203 | 703 | R>I | No | gnomAD | |
| rs1377141203 | 703 | R>K | No | gnomAD | |
| rs1331129132 | 704 | R>C | No | TOPMed | |
| rs1346946863 | 704 | R>H | No | gnomAD | |
| rs980267265 | 706 | A>T | No | Ensembl | |
| rs780740899 | 708 | H>R | No |
ExAC gnomAD |
|
| rs1277742301 | 708 | H>Y | No | TOPMed | |
| rs142716076 | 712 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs142716076 | 712 | V>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1415755018 | 713 | K>E | No |
TOPMed gnomAD |
|
| rs372673553 | 715 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1220474079 | 716 | K>R | No | Ensembl | |
| rs777857435 | 719 | V>L | No |
ExAC gnomAD |
|
| rs777857435 | 719 | V>M | No |
ExAC gnomAD |
|
| rs1223863994 | 720 | H>D | No |
TOPMed gnomAD |
|
| COSM4819151 | 720 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1893327834 | 720 | H>R | No | TOPMed | |
| rs1595477132 | 721 | D>G | No | gnomAD | |
| rs1204345458 | 721 | D>Y | No | Ensembl | |
| rs1158031130 | 725 | H>N | No | gnomAD | |
| rs1893326926 | 726 | Q>* | No | gnomAD | |
| rs1231736947 | 727 | L>V | No | gnomAD | |
| rs1468707634 | 728 | A>T | No | gnomAD | |
| rs1433482900 | 730 | Q>H | No | gnomAD | |
| rs2139459780 | 730 | Q>R | No | Ensembl | |
| TCGA novel | 732 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767221869 | 733 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs750228190 | 733 | I>S | No | ExAC | |
| rs1485218238 | 733 | I>V | No | gnomAD | |
| rs761471826 | 734 | L>V | No |
ExAC gnomAD |
|
| rs377340354 | 735 | M>T | No |
ESP ExAC gnomAD |
|
| rs768243510 | 737 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1893324796 | 740 | L>S | No | TOPMed | |
| rs1293512728 | 742 | K>E | No | gnomAD | |
| rs148812684 | 742 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1595477072 | 742 | K>T | No | Ensembl | |
| rs1893324039 | 743 | S>A | No | Ensembl | |
| rs373866056 | 745 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs373866056 | 745 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1430712932 | 746 | E>K | No | gnomAD | |
| rs369025395 | 747 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1334503033 | 748 | H>R | No |
TOPMed gnomAD |
|
| rs147781622 | 749 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs367908402 | 750 | E>K | No |
ESP TOPMed gnomAD |
|
| rs1011466308 | 751 | M>I | No | TOPMed | |
| rs764812294 | 751 | M>L | No |
ExAC gnomAD |
|
| rs2139459062 | 755 | V>A | No | Ensembl | |
| rs947392507 | 755 | V>I | No | Ensembl | |
| rs1440234999 | 756 | G>D | No |
TOPMed gnomAD |
|
| rs1219291529 | 756 | G>R | No |
TOPMed gnomAD |
|
| rs1219291529 | 756 | G>S | No |
TOPMed gnomAD |
|
| rs776230318 | 757 | T>K | No |
ExAC gnomAD |
|
| rs1354189397 | 758 | I>L | No | gnomAD | |
| rs1893313932 | 758 | I>M | No | TOPMed | |
| rs1354189397 | 758 | I>V | No | gnomAD | |
| rs1257219751 | 760 | D>N | No | TOPMed | |
| rs1257219751 | 760 | D>Y | No | TOPMed | |
| rs773562932 | 762 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs761250926 | 762 | Y>H | No |
ExAC gnomAD |
|
| rs1053399593 | 763 | E>K | No |
TOPMed gnomAD |
|
| rs762000637 | 764 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs747092497 | 765 | E>K | No |
ExAC gnomAD |
|
| rs2139458895 | 767 | A>P | No | Ensembl | |
|
COSM1740089 rs374402137 |
767 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1249287268 | 768 | M>T | No | gnomAD | |
| rs1480077851 | 768 | M>V | No | gnomAD | |
| rs747006250 | 771 | D>E | No |
ExAC gnomAD |
|
| rs552060222 | 771 | D>N | No | Ensembl | |
| rs1310959186 | 773 | N>S | No | gnomAD | |
| rs1893310790 | 775 | K>Q | No | TOPMed | |
| rs1309175294 | 775 | K>T | No | gnomAD | |
| rs1464333227 | 777 | T>A | No | gnomAD | |
| rs758312268 | 778 | A>G | No |
ExAC gnomAD |
|
| rs758312268 | 778 | A>V | No |
ExAC gnomAD |
|
| rs1380286515 | 780 | N>K | No | TOPMed | |
| rs948489921 | 780 | N>T | No | Ensembl | |
| rs747896976 | 783 | L>I | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 784 | C>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1893247232 | 785 | S>C | No | Ensembl | |
| rs754583657 | 786 | F>L | No |
ExAC gnomAD |
|
| rs753586356 | 787 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs371074129 | 788 | D>G | No |
ESP TOPMed gnomAD |
|
| rs1893246117 | 793 | Q>E | No | Ensembl | |
| rs1893246117 | 793 | Q>K | No | Ensembl | |
| rs749965334 | 796 | Q>R | No |
ExAC gnomAD |
|
| rs1485832467 | 798 | E>A | No | gnomAD | |
| rs202067243 | 799 | D>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1893245049 | 799 | D>N | No |
TOPMed gnomAD |
|
| rs202067243 | 799 | D>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1335667114 | 802 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| TCGA novel | 805 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1292947808 | 806 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs147925899 | 807 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs147925899 | 807 | K>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs764583553 | 808 | K>E | No |
ExAC gnomAD |
|
| rs1893243497 | 812 | A>S | No |
TOPMed gnomAD |
|
| rs1893243341 | 812 | A>V | No | TOPMed | |
| COSM3493944 | 814 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1386376247 | 814 | W>C | No | gnomAD | |
| rs763068962 | 814 | W>R | No |
ExAC gnomAD |
|
| rs1327284498 | 815 | E>K | No | gnomAD | |
| rs1566865374 | 816 | A>V | No | Ensembl | |
| rs1461545624 | 817 | Q>L | No | gnomAD | |
| rs1328868864 | 818 | I>M | No | gnomAD | |
| rs1161157122 | 818 | I>V | No | gnomAD | |
| rs377033153 | 819 | A>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs377033153 | 819 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1194368118 | 820 | E>G | No | gnomAD | |
| rs149815807 | 821 | I>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs149815807 | 821 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs868836765 | 823 | Q>* | No | TOPMed | |
| rs765766020 | 826 | S>N | No |
ExAC gnomAD |
|
| rs754162942 | 828 | E>K | No |
ExAC gnomAD |
|
| rs754162942 | 828 | E>Q | No |
ExAC gnomAD |
|
| rs766537181 | 829 | K>N | No |
ExAC gnomAD |
|
| TCGA novel | 830 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1595474588 | 831 | A>S | No | Ensembl | |
| rs1893201129 | 831 | A>V | No | gnomAD | |
| rs750491753 | 832 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs760767641 COSM4049235 |
832 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs890520524 | 833 | G>D | No | gnomAD | |
| rs890520524 | 833 | G>V | No | gnomAD | |
| rs1893200417 | 837 | A>V | No | TOPMed | |
| rs1893200172 | 840 | S>F | No | TOPMed | |
| rs762802616 | 841 | K>E | No |
ExAC gnomAD |
|
| rs762802616 | 841 | K>Q | No |
ExAC gnomAD |
|
| TCGA novel | 842 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2139451018 | 842 | M>V | No | Ensembl | |
| rs775258543 | 843 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs775258543 | 843 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1350831582 | 844 | E>K | No | gnomAD | |
| rs1393239915 | 845 | E>Q | No | Ensembl | |
| rs1253873946 | 846 | L>V | No |
TOPMed gnomAD |
|
| rs1179776450 | 847 | E>K | No |
TOPMed gnomAD |
|
| COSM3493943 | 847 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2139450879 | 849 | L>F | No | Ensembl | |
| rs1893198579 | 850 | R>K | No | Ensembl | |
| rs1893198451 | 850 | R>S | No | TOPMed | |
| rs1893198335 | 851 | S>C | No | TOPMed | |
| rs1226118771 | 851 | S>N | No | gnomAD | |
| rs776302347 | 852 | S>P | No |
ExAC gnomAD |
|
| rs189496293 | 853 | S>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1359074921 | 854 | L>R | No | gnomAD | |
| rs749560849 | 854 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1893197344 | 856 | S>L | No | gnomAD | |
| rs777154597 | 856 | S>T | No |
ExAC gnomAD |
|
| rs1893197099 | 857 | R>G | No | Ensembl | |
| rs373223628 | 858 | T>A | No |
ESP ExAC gnomAD |
|
| rs1158541124 | 859 | L>V | No | gnomAD | |
| rs1595472810 | 860 | D>A | No | Ensembl | |
| rs1319568762 | 861 | P>L | No |
TOPMed gnomAD |
|
| rs1319568762 | 861 | P>Q | No |
TOPMed gnomAD |
|
| rs1319568762 | 861 | P>R | No |
TOPMed gnomAD |
|
| rs1595472779 | 865 | V>G | No | Ensembl | |
| rs781307670 | 866 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2139444260 | 866 | R>H | No | 1000Genomes | |
| rs1893111026 | 870 | K>R | No | Ensembl | |
| rs1297582154 | 872 | D>G | No | gnomAD | |
| rs1373367735 | 875 | A>V | No |
TOPMed gnomAD |
|
| rs1361684372 | 878 | E>Q | No | TOPMed | |
| rs1283301759 | 881 | S>A | No | gnomAD | |
| rs1410362571 | 881 | S>L | No | gnomAD | |
| TCGA novel | 885 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4049233 rs368109666 |
885 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1234297974 | 888 | R>Q | No |
TOPMed gnomAD |
|
| rs1487279455 | 888 | R>W | No | gnomAD | |
| TCGA novel | 889 | A>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772783051 | 889 | A>T | No |
ExAC gnomAD |
|
| rs1343395143 | 892 | L>V | No | gnomAD | |
| TCGA novel | 893 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1226743911 | 894 | Q>R | No | gnomAD | |
| rs376060960 | 895 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1302009178 | 897 | L>V | No | gnomAD | |
| rs774010814 | 899 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1595472621 | 900 | V>G | No | Ensembl | |
| rs1369645777 | 900 | V>I | No | gnomAD | |
| rs1436455358 | 902 | D>A | No | gnomAD | |
| rs1305970504 | 902 | D>Y | No | gnomAD | |
| rs1893104812 | 903 | A>D | No | TOPMed | |
| rs142018795 | 903 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1893104681 | 904 | N>D | No | TOPMed | |
| rs200090522 | 904 | N>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM4925430 | 904 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200090522 | 904 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200090522 | 904 | N>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1893104003 | 906 | T>I | No | TOPMed | |
| rs2139443589 | 907 | L>F | No | Ensembl | |
| rs1893103610 | 907 | L>S | No | Ensembl | |
| rs1010112663 | 908 | E>G | No | Ensembl | |
| rs1893035908 | 909 | S>R | No | TOPMed | |
| rs1403264413 | 910 | K>E | No | gnomAD | |
| rs368505938 | 910 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1430397174 | 913 | D>H | No |
TOPMed gnomAD |
|
| rs1430397174 | 913 | D>N | No |
TOPMed gnomAD |
|
| rs1893035463 | 913 | D>V | No | Ensembl | |
| rs1430397174 | 913 | D>Y | No |
TOPMed gnomAD |
|
| rs1893035349 | 914 | S>F | No | TOPMed | |
| rs747293174 | 915 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1172202091 | 916 | A>D | No | gnomAD | |
| rs1172202091 | 916 | A>G | No | gnomAD | |
| COSM1368389 | 918 | N>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1209477360 | 919 | R>G | No |
TOPMed gnomAD |
|
| rs1383622394 | 919 | R>T | No |
TOPMed gnomAD |
|
| COSM4049232 | 921 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777811682 | 924 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs979915485 | 924 | E>K | No |
TOPMed gnomAD |
|
| rs1595471008 | 925 | M>R | No | Ensembl | |
| rs954036769 | 925 | M>V | No | Ensembl | |
| rs1363591237 | 927 | I>V | No | TOPMed | |
| rs1234895122 | 930 | K>Q | No |
TOPMed gnomAD |
|
| rs1340917277 | 932 | M>V | No |
TOPMed gnomAD |
|
| rs1893033371 | 933 | E>K | No | TOPMed | |
| rs779809375 | 934 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs756091661 | 935 | K>N | No |
ExAC gnomAD |
|
| rs1893032782 | 937 | R>G | No | TOPMed | |
| rs1011850433 | 938 | A>V | No | Ensembl | |
| COSM3814282 | 939 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1456288214 | 940 | T>I | No | gnomAD | |
| rs1456288214 | 940 | T>S | No | gnomAD | |
| rs1169851926 | 941 | G>E | No | gnomAD | |
| rs1246209355 | 942 | L>F | No |
TOPMed gnomAD |
|
| rs376599246 | 944 | L>I | No |
ESP TOPMed gnomAD |
|
| rs376599246 | 944 | L>V | No |
ESP TOPMed gnomAD |
|
| rs373398080 | 945 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1212482889 | 945 | P>R | No | TOPMed | |
| rs1179475383 | 946 | D>G | No | TOPMed | |
| rs764790988 | 946 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs764790988 | 946 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs777516650 | 948 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1159318563 | 949 | D>E | No |
TOPMed gnomAD |
|
| rs1469832334 | 949 | D>Y | No |
TOPMed gnomAD |
|
|
COSM3793488 rs1892868023 |
951 | I>V | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed gnomAD |
| TCGA novel | 953 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 953 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1414326490 | 956 | N>S | No | gnomAD | |
| rs200543279 | 957 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs764862437 | 959 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs759941973 | 959 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs764862437 | 959 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1040234745 | 960 | L>F | No |
TOPMed gnomAD |
|
| rs766903694 | 960 | L>P | No |
ExAC gnomAD |
|
| rs760979271 | 961 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs760979271 | 961 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs773581621 | 962 | H>R | No |
ExAC gnomAD |
|
| TCGA novel | 963 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1429625007 | 965 | T>K | No | gnomAD | |
| rs761918058 | 968 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs556232471 | 969 | S>N | No | gnomAD | |
| rs556232471 | 969 | S>T | No | gnomAD | |
|
rs760759588 TCGA novel |
973 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs760759588 | 973 | E>Q | No | Ensembl | |
| rs377345721 | 974 | Q>H | No |
ESP TOPMed |
|
| rs539199355 | 974 | Q>P | No |
TOPMed gnomAD |
|
| rs539199355 | 974 | Q>R | No |
TOPMed gnomAD |
|
| rs2139408093 | 975 | E>A | No | Ensembl | |
| rs1324585188 | 976 | T>I | No | TOPMed | |
| rs373015493 | 977 | Q>E | No |
ESP TOPMed |
|
| rs373015493 | 977 | Q>K | No |
ESP TOPMed |
|
| TCGA novel | 978 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774672648 | 979 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs764250909 | 981 | P>S | No |
ExAC gnomAD |
|
| rs763035580 | 982 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs142167570 | 983 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs142167570 | 983 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs771946677 | 984 | S>C | No |
ExAC gnomAD |
|
| rs771946677 | 984 | S>F | No |
ExAC gnomAD |
|
| rs547015130 | 985 | P>A | No |
TOPMed gnomAD |
|
| rs201476818 | 985 | P>L | No |
1000Genomes gnomAD |
|
| rs547015130 | 985 | P>S | No |
TOPMed gnomAD |
|
| rs1158789055 | 986 | S>L | No | gnomAD | |
| rs1445844152 | 987 | M>I | No | gnomAD | |
| rs754600095 | 987 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1415616131 | 987 | M>V | No | gnomAD | |
| rs1892634865 | 988 | S>A | No | TOPMed | |
|
rs200096986 COSM953599 |
988 | S>F | endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1892634443 | 989 | V>A | No | TOPMed | |
| rs1456240485 | 989 | V>L | No |
TOPMed gnomAD |
|
| rs1456240485 | 989 | V>M | No |
TOPMed gnomAD |
|
| rs779607293 | 991 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs750975756 | 992 | S>P | No |
ExAC gnomAD |
|
| TCGA novel | 993 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1247029156 | 994 | Q>H | No |
TOPMed gnomAD |
|
| rs1161351674 | 995 | Q>R | No |
TOPMed gnomAD |
|
| rs1595462690 | 996 | E>G | No | Ensembl | |
| rs769333187 | 997 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs374316100 | 998 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs781685946 | 1000 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs781685946 | 1000 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs960423641 | 1000 | R>W | No |
TOPMed gnomAD |
|
| rs186167635 | 1002 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1166146982 | 1003 | Q>* | No | gnomAD | |
| rs1892618320 | 1003 | Q>R | No | TOPMed | |
| rs2139406425 | 1004 | R>T | No | Ensembl | |
| rs962655009 | 1007 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1190204656 | 1007 | A>V | No | gnomAD | |
| rs1476842864 | 1008 | V>M | No |
TOPMed gnomAD |
|
| rs1016858776 | 1009 | P>L | No |
TOPMed gnomAD |
|
| rs1207773726 | 1009 | P>T | No | gnomAD | |
| rs1202514739 | 1011 | P>L | No | gnomAD | |
| rs1218811841 | 1013 | T>M | No |
TOPMed gnomAD |
|
| rs1343898761 | 1014 | Q>R | No | gnomAD | |
| rs1289940468 | 1015 | A>S | No | gnomAD | |
| rs1306175978 | 1017 | A>P | No |
TOPMed gnomAD |
|
| rs1306175978 | 1017 | A>S | No |
TOPMed gnomAD |
|
| rs868119630 | 1019 | A>V | No | Ensembl | |
| rs1026416001 | 1021 | P>L | No |
TOPMed gnomAD |
|
| rs1026416001 | 1021 | P>R | No |
TOPMed gnomAD |
|
| rs1892539787 | 1023 | P>L | No | TOPMed | |
| rs1181332127 | 1024 | K>E | No | gnomAD | |
| rs374839429 | 1025 | A>P | No |
ESP ExAC gnomAD |
|
| rs374839429 | 1025 | A>T | No |
ESP ExAC gnomAD |
|
| TCGA novel | 1025 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs941942131 | 1028 | F>L | No |
TOPMed gnomAD |
|
| rs1892539175 | 1029 | S>G | No | gnomAD | |
| rs1892539175 | 1029 | S>R | No | gnomAD | |
| rs1261983545 | 1030 | I>L | No | TOPMed | |
| TCGA novel | 1030 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1249983880 | 1031 | K>R | No | gnomAD | |
| rs1892538665 | 1034 | S>A | No | Ensembl | |
| rs1209532969 | 1035 | S>T | No | gnomAD | |
| rs1595460747 | 1039 | C>S | No | Ensembl | |
| rs1048432907 | 1040 | S>N | No | Ensembl | |
| rs1048432907 | 1040 | S>T | No | Ensembl | |
| rs1258692571 | 1041 | H>Y | No | gnomAD | |
| rs372111401 | 1044 | S>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1892537262 | 1048 | G>E | No | gnomAD | |
| rs61733950 | 1049 | L>V | No | gnomAD | |
| rs755257364 | 1051 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs755257364 | 1051 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs766429486 | 1054 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs751490803 | 1055 | A>T | No |
ExAC TOPMed gnomAD |
|
| COSM3493942 | 1055 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs367927706 | 1057 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs963853088 | 1058 | V>A | No | Ensembl | |
| rs1595458911 | 1060 | S>T | No | Ensembl | |
| COSM697011 | 1065 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1220016248 | 1065 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| COSM3793487 | 1066 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1067 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs140416985 | 1069 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs758332208 | 1070 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1336802752 | 1070 | G>D | No |
TOPMed gnomAD |
|
| rs758332208 | 1070 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs752480463 | 1071 | A>T | No |
ExAC gnomAD |
|
| rs765072170 | 1071 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs759201646 | 1073 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs1283001163 | 1073 | Q>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1368387 | 1073 | Q>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776110379 | 1074 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1004613613 | 1076 | P>L | No | Ensembl | |
|
VAR_025847 rs34822377 |
1077 | I>V | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1892440883 | 1078 | P>L | No | Ensembl | |
| rs1892441005 | 1078 | P>S | No | Ensembl | |
| rs1029990072 | 1079 | P>A | No |
TOPMed gnomAD |
|
| rs1892440297 | 1080 | E>G | No | gnomAD | |
| rs772702155 | 1080 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs772702155 | 1080 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs748723581 | 1081 | Q>P | No |
ExAC gnomAD |
|
| rs1466712824 | 1083 | K>E | No |
TOPMed gnomAD |
|
| rs1466712824 | 1083 | K>Q | No |
TOPMed gnomAD |
|
| rs774836834 | 1084 | R>S | No |
ExAC gnomAD |
|
| rs765867653 | 1085 | P>A | No |
TOPMed gnomAD |
|
| rs780088812 | 1088 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs780088812 | 1088 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs376059761 | 1089 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1200571246 | 1090 | V>M | No |
TOPMed gnomAD |
|
|
COSM20501 rs1260927406 |
1092 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs946145684 | 1092 | R>Q | No | TOPMed | |
| rs758315719 | 1094 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs776378296 | 1094 | I>T | No | ExAC | |
| rs1289391194 | 1095 | G>R | No | gnomAD | |
| rs1355936852 | 1096 | T>A | No | TOPMed | |
| rs1445276916 | 1097 | A>V | No | TOPMed | |
| rs1301772163 | 1098 | Y>H | No | TOPMed | |
| rs1334546462 | 1100 | G>S | No | gnomAD | |
| TCGA novel | 1103 | K>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs571661306 | 1104 | V>I | No | Ensembl | |
| rs1487347096 | 1107 | P>A | No |
TOPMed gnomAD |
|
| rs1892404106 | 1108 | T>A | No | gnomAD | |
| TCGA novel | 1108 | T>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759721687 | 1108 | T>M | No |
ExAC gnomAD |
|
| rs1384300410 | 1109 | G>E | No |
TOPMed gnomAD |
|
| rs1595457972 | 1110 | V>G | No | Ensembl | |
| rs1281732707 | 1116 | R>C | No |
TOPMed gnomAD |
|
| rs1428890014 | 1116 | R>H | No | gnomAD | |
| rs142564408 | 1117 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1892402177 | 1118 | Y>F | No | TOPMed | |
| rs768469311 | 1119 | A>G | No |
ExAC gnomAD |
|
| TCGA novel | 1119 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768469311 | 1119 | A>V | No |
ExAC gnomAD |
|
| rs779685101 | 1121 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs779685101 | 1121 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1039611600 | 1123 | D>H | No | gnomAD | |
| rs755934852 | 1125 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1248777092 | 1126 | L>I | No |
TOPMed gnomAD |
|
| rs1892400139 | 1129 | Y>* | No | gnomAD | |
| TCGA novel | 1129 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1490323123 | 1129 | Y>H | No | gnomAD | |
| rs943515361 | 1130 | D>H | No | Ensembl | |
| TCGA novel | 1130 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780931165 | 1131 | L>P | No | ExAC | |
| rs1595457897 | 1137 | T>P | No | Ensembl | |
| rs1892399212 | 1139 | P>S | No | gnomAD | |
| rs1267828637 | 1142 | I>N | No | TOPMed | |
| rs1267828637 | 1142 | I>T | No | TOPMed | |
| rs756971709 | 1142 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1277468665 | 1143 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1892397974 | 1144 | S>N | No |
TOPMed gnomAD |
|
| COSM697013 | 1149 | L>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763345731 | 1149 | L>F | No |
ExAC gnomAD |
|
| rs1359377150 | 1150 | R>S | No | gnomAD | |
| rs1465024027 | 1151 | D>G | No |
TOPMed gnomAD |
|
| rs150060973 | 1152 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC TOPMed gnomAD NCI-TCGA |
| rs539521711 | 1153 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1354986103 | 1155 | S>F | No | TOPMed | |
| rs1420176623 | 1161 | A>V | No |
TOPMed gnomAD |
|
| rs1052494408 | 1163 | D>G | No | Ensembl | |
| rs1892249517 | 1164 | V>I | No | Ensembl | |
| rs201174859 | 1168 | T>A | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs769846883 | 1169 | R>C | No |
ExAC gnomAD |
|
| rs547906994 | 1169 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs547906994 | 1169 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM1368385 | 1169 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1269749428 | 1170 | R>Q | No |
TOPMed gnomAD |
|
| rs1239562219 | 1171 | D>N | No | gnomAD | |
|
TCGA novel rs1892248315 |
1172 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1279171271 | 1174 | C>S | No |
TOPMed gnomAD |
|
| rs369632217 | 1179 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
|
rs369632217 COSM4049230 |
1179 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1293777219 | 1181 | S>C | No |
TOPMed gnomAD |
|
| rs1293777219 | 1181 | S>F | No |
TOPMed gnomAD |
|
| rs1464547211 | 1182 | L>F | No | gnomAD | |
| rs1892192242 | 1183 | L>F | No | TOPMed | |
| rs1892192110 | 1184 | G>C | No | gnomAD | |
|
RCV001814852 rs1892191977 |
1184 | G>D | No |
ClinVar TOPMed dbSNP |
|
| rs1259349822 | 1185 | A>E | No |
TOPMed gnomAD |
|
| rs1259349822 | 1185 | A>G | No |
TOPMed gnomAD |
|
| rs1030181382 | 1186 | P>S | No | TOPMed | |
| rs781464184 | 1190 | S>C | No |
ExAC gnomAD |
|
| rs781464184 | 1190 | S>G | No |
ExAC gnomAD |
|
| rs1566846568 | 1190 | S>V | No | Ensembl | |
| rs1291023466 | 1191 | S>L | No | gnomAD | |
| COSM3814281 | 1202 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
VAR_070886 rs146298297 |
1203 | R>K | No |
UniProt ESP ExAC dbSNP gnomAD |
|
| rs368543743 | 1204 | K>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1407202788 | 1204 | K>R | No | gnomAD | |
| rs1346688931 | 1205 | W>C | No | gnomAD | |
| rs766338362 | 1206 | V>I | No |
ExAC TOPMed gnomAD |
|
| COSM953597 | 1209 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs773038838 | 1213 | Q>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1215 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs771803261 | 1216 | L>F | No |
ExAC gnomAD |
|
| rs1187897758 | 1217 | H>L | No | TOPMed | |
| rs1461641399 | 1217 | H>Y | No |
TOPMed gnomAD |
|
| rs771945943 | 1220 | R>G | No | gnomAD | |
| rs538525924 | 1220 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs771945943 | 1220 | R>W | No | gnomAD | |
| rs773947478 | 1221 | L>R | No |
ExAC gnomAD |
|
| rs748732527 | 1223 | N>S | No |
ExAC gnomAD |
|
| rs1250571758 | 1224 | Q>R | No | gnomAD | |
| rs779603324 | 1225 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs934380429 | 1226 | V>L | No |
TOPMed gnomAD |
|
| rs934380429 | 1226 | V>M | No |
TOPMed gnomAD |
|
| rs746406995 | 1227 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1892185442 | 1228 | V>I | No | TOPMed | |
| TCGA novel | 1229 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747398820 | 1234 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1311063319 | 1235 | S>N | No | gnomAD | |
| rs1892184656 | 1235 | S>R | No | gnomAD | |
| rs1342134723 | 1236 | S>L | No |
TOPMed gnomAD |
|
| rs758761562 | 1238 | P>H | No |
ExAC gnomAD |
|
|
rs765283166 COSM3493940 |
1239 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs765283166 | 1239 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs1428457211 | 1240 | I>L | No | TOPMed | |
| COSM953596 | 1245 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756048171 | 1246 | A>V | No |
ExAC gnomAD |
|
| rs1471104141 | 1247 | A>D | No | gnomAD | |
| COSM953595 | 1248 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767517705 | 1248 | I>T | No |
ExAC gnomAD |
|
| rs1892182282 | 1248 | I>V | No | gnomAD | |
| rs774139239 | 1249 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1216614824 | 1251 | A>G | No |
TOPMed gnomAD |
|
| rs1449911639 | 1251 | A>T | No | Ensembl | |
| rs748549609 | 1252 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1245100358 | 1255 | A>T | No | TOPMed | |
| rs1293446771 | 1256 | V>A | No |
TOPMed gnomAD |
|
| rs1306608589 | 1256 | V>F | No |
TOPMed gnomAD |
|
| rs1203790110 | 1257 | G>S | No | TOPMed | |
| rs202026444 | 1262 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs202026444 | 1262 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751534717 | 1263 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs777715596 | 1265 | I>L | No |
ExAC gnomAD |
|
| rs1423522944 | 1266 | E>Q | No |
TOPMed gnomAD |
|
| rs1382859316 | 1268 | T>P | No | gnomAD | |
| rs140036206 | 1269 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs752549385 | 1269 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs752549385 | 1269 | R>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1270 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1892057228 | 1271 | V>A | No | Ensembl | |
| rs770015832 | 1272 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1393940175 | 1273 | V>A | No |
TOPMed gnomAD |
|
| rs745881611 | 1273 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs1393940175 | 1273 | V>G | No |
TOPMed gnomAD |
|
| rs745881611 | 1273 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1202945188 | 1274 | R>C | No |
TOPMed gnomAD |
|
| rs1595450511 | 1274 | R>H | No | Ensembl | |
| rs150629546 | 1276 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1277 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1454734405 | 1277 | D>Y | No | gnomAD | |
| rs778749230 | 1278 | C>F | No |
ExAC TOPMed gnomAD |
|
| rs778749230 | 1278 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs778749230 | 1278 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs754754131 | 1281 | V>L | No |
1000Genomes ExAC gnomAD |
|
| rs779790085 | 1282 | H>Q | No |
ExAC gnomAD |
|
| rs749051826 | 1282 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1595450479 | 1283 | Q>* | No | Ensembl | |
| rs1385950616 | 1283 | Q>R | No | gnomAD | |
| rs1321818416 | 1284 | I>F | No |
TOPMed gnomAD |
|
| rs749941853 | 1285 | E>G | No | ExAC | |
|
rs764023925 COSM239242 |
1285 | E>K | Variant assessed as Somatic; MODERATE impact. prostate [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1595450440 | 1288 | P>L | No | TOPMed | |
| rs1485493716 | 1288 | P>S | No | gnomAD | |
| rs752062947 | 1289 | R>K | No |
ExAC gnomAD |
|
| TCGA novel | 1290 | E>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1892052545 | 1291 | K>E | No | Ensembl | |
| rs1249602705 | 1291 | K>N | No | Ensembl | |
| rs1480011161 | 1292 | I>M | No |
TOPMed gnomAD |
|
| rs763316431 | 1292 | I>T | No |
ExAC gnomAD |
|
| rs1260603179 | 1292 | I>V | No |
TOPMed gnomAD |
|
| rs775835433 | 1293 | V>I | No |
ExAC TOPMed gnomAD |
|
| COSM4049229 | 1294 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765175299 | 1295 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs866343922 | 1296 | L>F | No | Ensembl | |
| rs759811615 | 1299 | R>W | No |
ExAC gnomAD |
|
| rs1268497380 | 1300 | N>H | No | gnomAD | |
| rs1340448395 | 1300 | N>S | No |
TOPMed gnomAD |
|
| rs1407316427 | 1303 | V>A | No | gnomAD | |
| rs771002666 | 1304 | H>Q | No |
ExAC gnomAD |
|
| rs1034656188 | 1304 | H>R | No | Ensembl | |
| rs1364251429 | 1304 | H>Y | No |
TOPMed gnomAD |
|
| rs1162776652 | 1305 | L>V | No | gnomAD | |
|
COSM1235984 rs905113104 |
1306 | Y>C | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1375056196 | 1307 | P>A | No | gnomAD | |
| rs1341355633 | 1307 | P>L | No | gnomAD | |
| rs1375056196 | 1307 | P>S | No | gnomAD | |
| rs1427632487 | 1309 | S>L | No |
TOPMed gnomAD |
|
| rs1278543765 | 1310 | S>A | No | TOPMed | |
| rs1595450310 | 1312 | D>V | No | Ensembl | |
| rs749094142 | 1314 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs749094142 | 1314 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs749094142 | 1314 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| VAR_040838 | 1315 | E>K | a lung large cell carcinoma sample; somatic mutation [UniProt] | No | UniProt |
| rs1566843791 | 1317 | S>C | No | Ensembl | |
| rs1226248828 | 1317 | S>I | No |
TOPMed gnomAD |
|
| rs1226248828 | 1317 | S>T | No |
TOPMed gnomAD |
|
| rs1892046481 | 1322 | L>V | No |
TOPMed gnomAD |
|
| rs201120777 | 1323 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201120777 | 1323 | P>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM4492348 rs1566843777 |
1323 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs764593988 | 1326 | K>R | No |
ExAC gnomAD |
|
| rs1465064771 | 1327 | G>S | No | gnomAD | |
| rs1892045402 | 1328 | C>S | No | gnomAD | |
|
rs2139363710 RCV001390771 |
1329 | Q>* | No |
ClinVar Ensembl dbSNP |
|
| rs758892375 | 1330 | L>F | No |
ExAC gnomAD |
|
| rs753046475 | 1331 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs753046475 | 1331 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1595450226 | 1332 | A>P | No | Ensembl | |
|
COSM1166004 rs539439670 |
1333 | T>M | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1595450223 | 1333 | T>P | No | Ensembl | |
| rs776785413 | 1334 | A>V | No |
ExAC gnomAD |
|
| rs766525858 | 1335 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs766525858 | 1335 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs2139363582 | 1336 | L>R | No | Ensembl | |
| rs767751908 | 1337 | K>E | No |
ExAC TOPMed gnomAD |
|
|
COSM953593 rs767751908 |
1337 | K>Q | endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs768628164 | 1337 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1892043395 | 1338 | R>K | No | Ensembl | |
| rs1393359065 | 1340 | S>P | No | TOPMed | |
| rs763037888 | 1341 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs759807460 | 1342 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs775397689 | 1342 | T>P | No |
ExAC gnomAD |
|
| rs775397689 | 1342 | T>S | No |
ExAC gnomAD |
|
| rs745676686 | 1343 | C>F | No |
ExAC gnomAD |
|
| rs1164558007 | 1345 | F>L | No | gnomAD | |
| rs1324880466 | 1348 | V>A | No | gnomAD | |
| rs1324880466 | 1348 | V>G | No | gnomAD | |
|
COSM4049228 rs746522635 |
1348 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs777205662 | 1350 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs143564466 | 1351 | L>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs753163105 | 1352 | I>M | No |
ExAC gnomAD |
|
| rs1595450117 | 1352 | I>V | No | Ensembl | |
| rs1398437447 | 1353 | L>F | No | gnomAD | |
| rs2139363277 | 1355 | Y>* | No | Ensembl | |
| rs755279217 | 1355 | Y>C | No |
ExAC gnomAD |
|
| rs1401243402 | 1358 | Q>E | No | gnomAD | |
| rs1892039555 | 1358 | Q>H | No | TOPMed | |
| rs1170684714 | 1358 | Q>R | No | gnomAD | |
| rs754160186 | 1359 | R>K | No |
ExAC gnomAD |
|
| rs528394229 | 1360 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1892038948 | 1361 | K>R | No | TOPMed | |
| rs1892038662 | 1366 | K>R | No | Ensembl | |
| rs1892038172 | 1368 | N>K | No | Ensembl | |
| rs199903511 | 1368 | N>S | No |
1000Genomes TOPMed gnomAD |
|
| rs1437416700 | 1368 | N>Y | No | gnomAD | |
| rs1194977739 | 1371 | V>L | No |
TOPMed gnomAD |
|
| rs1892037577 | 1372 | A>V | No | TOPMed | |
| rs1892037422 | 1373 | P>L | No | Ensembl | |
| rs767538657 | 1374 | G>R | No |
ExAC gnomAD |
|
| rs767538657 | 1374 | G>S | No |
ExAC gnomAD |
|
| rs762012281 | 1375 | S>R | No |
ExAC gnomAD |
|
| rs774671721 | 1376 | V>M | No | Ensembl | |
| rs149319188 | 1379 | L>V | No |
ESP TOPMed gnomAD |
|
| rs1002396118 | 1380 | A>S | No | TOPMed | |
| rs1002396118 | 1380 | A>T | No | TOPMed | |
| rs759334009 | 1380 | A>V | No |
ExAC gnomAD |
|
| rs1595450021 | 1381 | V>G | No | Ensembl | |
| TCGA novel | 1383 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770601956 | 1384 | D>E | No |
ExAC TOPMed |
|
| rs1370672003 | 1384 | D>N | No | Ensembl | |
| rs746689190 | 1387 | C>F | No |
ExAC gnomAD |
|
| rs1167686182 | 1388 | V>A | No |
TOPMed gnomAD |
|
| rs777304874 | 1390 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs371972643 | 1390 | Y>H | No |
ESP TOPMed gnomAD |
|
| rs777304874 | 1390 | Y>S | No |
ExAC TOPMed gnomAD |
|
| rs529138453 | 1391 | P>L | No |
1000Genomes ExAC gnomAD |
|
| rs1595449952 | 1394 | F>V | No | Ensembl | |
| rs528455993 | 1395 | C>W | No | gnomAD | |
| rs1892032285 | 1397 | L>M | No | Ensembl | |
| rs755548130 | 1398 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1595449940 | 1399 | I>M | No | Ensembl | |
| rs1566843363 | 1400 | Q>L | No | Ensembl | |
| rs1566843363 | 1400 | Q>R | No | Ensembl | |
| COSM5940426 | 1401 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1334239878 | 1401 | G>R | No |
TOPMed gnomAD |
|
| rs548093157 | 1403 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs141912208 | 1404 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs141912208 | 1404 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1892030808 | 1406 | L>V | No | TOPMed | |
| rs1892030505 | 1407 | N>D | No | Ensembl | |
| rs1264618746 | 1409 | V>I | No | gnomAD | |
| rs145737227 | 1411 | P>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs761922472 | 1411 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs761922472 | 1411 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs145737227 | 1411 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1294074599 | 1412 | N>D | No |
TOPMed gnomAD |
|
| rs1294074599 | 1412 | N>H | No |
TOPMed gnomAD |
|
| rs1892029266 | 1412 | N>K | No | Ensembl | |
| rs751692127 | 1412 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs751692127 | 1412 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs1355965084 | 1414 | P>L | No | gnomAD | |
| rs1282194155 | 1415 | S>T | No | gnomAD | |
| rs1892028717 | 1415 | S>W | No | Ensembl | |
| rs1326806291 | 1416 | L>F | No |
TOPMed gnomAD |
|
| rs1892028040 | 1417 | A>T | No | TOPMed | |
|
COSM1368382 rs776462657 |
1417 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1163709689 | 1419 | L>F | No | gnomAD | |
| rs1427271427 | 1423 | S>C | No |
TOPMed gnomAD |
|
| rs773912384 | 1424 | F>S | No |
ExAC gnomAD |
|
| rs140245177 | 1425 | D>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs140245177 | 1425 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1271109107 | 1427 | L>R | No | gnomAD | |
| rs780645988 | 1431 | E>D | No |
ExAC gnomAD |
|
|
rs1287907520 COSM953591 |
1433 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1287907520 | 1433 | E>Q | No | gnomAD | |
| rs756382043 | 1434 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs151083985 | 1434 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1277362736 | 1435 | E>K | No |
TOPMed gnomAD |
|
| COSM3786213 | 1437 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1347373976 | 1440 | C>* | No | gnomAD | |
| rs1566843151 | 1443 | H>N | No | Ensembl | |
| rs1566843151 | 1443 | H>Y | No | Ensembl | |
| rs944799076 | 1444 | M>V | No |
TOPMed gnomAD |
|
| rs1892022642 | 1448 | V>M | No | Ensembl | |
| rs766354752 | 1450 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1477118006 | 1451 | Q>R | No | gnomAD | |
| rs773059863 | 1453 | R>G | No |
ExAC gnomAD |
|
| rs577521745 | 1453 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs773059863 | 1453 | R>W | No |
ExAC gnomAD |
|
| rs1892020736 | 1455 | A>S | No | Ensembl | |
| rs1206839670 | 1456 | R>C | No | gnomAD | |
| rs1458414574 | 1456 | R>H | No |
TOPMed gnomAD |
|
| rs1221432217 | 1457 | A>G | No |
TOPMed gnomAD |
|
|
rs564119735 COSM432695 |
1457 | A>T | breast [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1221432217 | 1457 | A>V | No |
TOPMed gnomAD |
|
| rs1892019501 | 1458 | Q>L | No | gnomAD | |
| rs1892019355 | 1459 | E>D | No | gnomAD | |
| rs775799497 | 1461 | M>I | No |
ExAC gnomAD |
|
| rs1892019190 | 1461 | M>L | No | TOPMed | |
| rs770334280 | 1463 | P>S | No |
ExAC gnomAD |
|
| rs763306847 | 1465 | A>G | No |
ExAC gnomAD |
|
| rs763306847 | 1465 | A>V | No |
ExAC gnomAD |
|
| rs140674190 | 1466 | P>S | No |
ESP ExAC |
|
| rs956441357 | 1467 | V>I | No | Ensembl | |
| rs546461837 | 1468 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs546461837 | 1468 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1408771029 | 1469 | C>F | No |
TOPMed gnomAD |
|
| rs1408771029 | 1469 | C>Y | No |
TOPMed gnomAD |
|
| rs771381267 | 1473 | P>L | No |
ExAC gnomAD |
|
| rs747400742 | 1474 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1313865039 | 1474 | T>I | No |
TOPMed gnomAD |
|
| rs747400742 | 1474 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs903725198 | 1475 | H>Y | No | Ensembl | |
| rs146870461 | 1476 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146870461 | 1476 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs748164773 | 1477 | T>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1479 | Y>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1048630181 | 1480 | S>R | No |
TOPMed gnomAD |
|
| rs1276345638 | 1481 | E>K | No |
TOPMed gnomAD |
|
| rs1426764810 | 1481 | E>V | No | gnomAD | |
| rs1359290677 | 1484 | V>L | No |
TOPMed gnomAD |
|
| rs1359290677 | 1484 | V>M | No |
TOPMed gnomAD |
|
| COSM4837716 | 1485 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2139347297 | 1485 | D>G | No | Ensembl | |
| rs61745673 | 1485 | D>N | No | Ensembl | |
| rs1421900220 | 1486 | V>I | No |
TOPMed gnomAD |
|
| rs1194354927 | 1490 | R>C | No | gnomAD | |
| rs1487900395 | 1490 | R>H | No | gnomAD | |
| rs1260441132 | 1491 | T>A | No | gnomAD | |
| rs1488606199 | 1492 | M>I | No | gnomAD | |
| rs1264559702 | 1495 | V>M | No | gnomAD | |
| TCGA novel | 1496 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1891831723 | 1497 | T>A | No |
TOPMed gnomAD |
|
| rs1320343582 | 1497 | T>S | No |
TOPMed gnomAD |
|
| rs376224473 | 1499 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs972830251 | 1501 | R>Q | No |
TOPMed gnomAD |
|
| rs529283722 | 1501 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1402233925 | 1502 | R>K | No | gnomAD | |
| rs773420791 | 1503 | I>V | No |
ExAC gnomAD |
|
| rs2139346627 | 1505 | P>T | No | Ensembl | |
| rs1891824675 | 1507 | N>D | No | TOPMed | |
|
rs1891824675 COSM226304 |
1507 | N>H | NS [Cosmic] | No |
cosmic curated TOPMed |
| rs774578569 | 1507 | N>S | No |
ExAC gnomAD |
|
| rs868573029 | 1508 | S>F | No | Ensembl | |
| rs368931708 | 1510 | G>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs368931708 | 1510 | G>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs749362454 | 1512 | L>F | No |
ExAC gnomAD |
|
| rs1247603618 | 1513 | N>S | No | TOPMed | |
| rs1247603618 | 1513 | N>T | No | TOPMed | |
| rs1891823520 | 1514 | L>H | No | gnomAD | |
| rs775600626 | 1515 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs770829901 | 1516 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs770829901 | 1516 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs201876066 | 1517 | C>* | No |
ExAC gnomAD |
|
| rs1891822723 | 1517 | C>F | No | Ensembl | |
| rs1891822816 | 1517 | C>R | No | TOPMed | |
| rs903757678 | 1518 | E>G | No | Ensembl | |
| rs1891822438 | 1518 | E>K | No |
TOPMed gnomAD |
|
| rs777601390 | 1519 | P>T | No |
ExAC TOPMed gnomAD |
|
|
rs747908716 COSM1188912 |
1521 | R>C | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs778561558 | 1521 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs747908716 | 1521 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs138717678 | 1524 | Y>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1380358092 | 1525 | F>L | No | gnomAD | |
|
rs750897008 TCGA novel |
1529 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC gnomAD |
| rs200886335 | 1530 | S>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767925910 | 1530 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1891820589 | 1531 | G>R | No | TOPMed | |
| rs202016085 | 1532 | A>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs749106487 | 1532 | A>T | No |
ExAC gnomAD |
|
| rs202016085 | 1532 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs745374413 | 1535 | N>S | No |
ExAC gnomAD |
|
| rs202209179 | 1536 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs202209179 | 1536 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2139345742 | 1537 | P>A | No | Ensembl | |
| rs752024361 | 1537 | P>L | No |
ExAC gnomAD |
|
| rs752024361 | 1537 | P>R | No |
ExAC gnomAD |
|
| rs758705817 | 1538 | D>G | No |
ExAC gnomAD |
|
| rs1200225915 | 1538 | D>H | No | gnomAD | |
| rs1216698851 | 1539 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs765442114 | 1539 | T>I | No |
ExAC gnomAD |
|
| rs759726532 | 1540 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs1474360437 | 1540 | S>P | No | Ensembl | |
| rs760828996 | 1541 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs766329158 | 1541 | D>G | No |
ExAC gnomAD |
|
| rs1009900223 | 1541 | D>N | No |
TOPMed gnomAD |
|
| rs544013491 | 1542 | N>D | No | gnomAD | |
| rs1310084124 | 1543 | S>C | No |
TOPMed gnomAD |
|
| rs774370821 | 1543 | S>N | No |
ExAC gnomAD |
|
| rs2139345596 | 1544 | K>R | No | Ensembl | |
| rs1427386882 | 1546 | Q>H | No | gnomAD | |
| rs762832845 | 1547 | M>I | No |
ExAC gnomAD |
|
| rs768715462 | 1547 | M>V | No |
ExAC gnomAD |
|
| rs1400235479 | 1548 | L>V | No |
TOPMed gnomAD |
|
| rs1449283610 | 1549 | R>C | No | gnomAD | |
|
COSM1732750 rs1251445406 |
1549 | R>H | pancreas [Cosmic] | No |
cosmic curated gnomAD |
| rs1251445406 | 1549 | R>P | No | gnomAD | |
| rs775479309 | 1550 | T>A | No |
ExAC gnomAD |
|
| rs1395543529 | 1550 | T>S | No | TOPMed | |
| rs1019328882 | 1551 | R>S | No |
TOPMed gnomAD |
|
| rs1891810515 | 1552 | S>T | No | TOPMed | |
| rs1335094844 | 1554 | R>S | No |
TOPMed gnomAD |
|
| rs373755648 | 1555 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1891810185 | 1555 | R>W | No | Ensembl | |
| rs1218057718 | 1557 | V>A | No | gnomAD | |
| rs780932163 | 1557 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1891809454 | 1559 | K>E | No | TOPMed | |
| rs770300795 | 1559 | K>R | No |
ExAC gnomAD |
|
|
rs1891809128 COSM697016 |
1562 | E>K | lung [Cosmic] | No |
cosmic curated TOPMed |
| rs2139345371 | 1563 | E>G | No | Ensembl | |
| COSM697018 | 1564 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1891808684 | 1568 | Q>H | No | TOPMed | |
|
rs1416279074 COSM1368380 |
1570 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs140004721 | 1570 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1313393417 | 1572 | M>T | No | gnomAD | |
| rs1414220354 | 1572 | M>V | No | gnomAD | |
| rs1191552046 | 1574 | R>K | No | TOPMed | |
| rs139323146 | 1575 | D>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1264728476 | 1579 | R>K | No | gnomAD | |
| rs1891801585 | 1579 | R>S | No | Ensembl | |
| TCGA novel | 1581 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1566839125 | 1581 | K>R | No | TOPMed | |
| rs1432873707 | 1582 | M>V | No |
TOPMed gnomAD |
|
| rs978643502 | 1585 | N>S | No |
TOPMed gnomAD |
|
| rs1379502143 | 1587 | T>I | No |
TOPMed gnomAD |
|
| rs1216621400 | 1592 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1351709514 | 1593 | A>V | No | gnomAD | |
| rs767750407 | 1595 | M>V | No |
ExAC TOPMed gnomAD |
|
| COSM3814279 | 1598 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757447340 | 1599 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1323922137 | 1600 | G>S | No |
TOPMed gnomAD |
|
|
COSM1514837 rs1313835255 COSM6139914 |
1601 | M>I | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated TOPMed gnomAD NCI-TCGA Cosmic |
| rs1459972650 | 1602 | Q>R | No | Ensembl | |
| rs1891799572 | 1603 | V>L | No | TOPMed | |
| rs759189098 | 1606 | D>A | No |
ExAC gnomAD |
|
| rs765236814 | 1606 | D>H | No |
ExAC gnomAD |
|
| rs765236814 | 1606 | D>Y | No |
ExAC gnomAD |
|
| rs763551730 | 1610 | S>I | No |
ExAC gnomAD |
|
| rs1339244595 | 1611 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1249699952 | 1612 | V>M | No | gnomAD | |
| rs1469719693 | 1613 | P>H | No |
TOPMed gnomAD |
|
| rs1469719693 | 1613 | P>L | No |
TOPMed gnomAD |
|
| rs142790424 | 1614 | P>A | No |
ESP ExAC gnomAD |
|
| rs781501373 | 1614 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs781501373 | 1614 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs781501373 | 1614 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1280778797 | 1615 | S>C | No |
TOPMed gnomAD |
|
| rs1280778797 | 1615 | S>F | No |
TOPMed gnomAD |
|
| rs781316314 | 1615 | S>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1595443617 | 1615 | S>P | No | Ensembl | |
| rs1280778797 | 1615 | S>Y | No |
TOPMed gnomAD |
|
| rs546966091 | 1616 | Q>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777958516 | 1618 | E>G | No |
ExAC gnomAD |
|
| rs138830696 | 1620 | P>L | No |
ESP TOPMed gnomAD |
|
| COSM4049227 | 1620 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs562034984 | 1621 | G>C | No |
1000Genomes ExAC gnomAD |
|
| rs1595443584 | 1621 | G>D | No | Ensembl | |
| rs1566838020 | 1622 | P>A | No | Ensembl | |
| rs755838432 | 1622 | P>L | No |
ExAC gnomAD |
|
| rs148797004 | 1623 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs148797004 | 1623 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1891735111 | 1624 | P>R | No | Ensembl | |
| rs750999657 | 1624 | P>T | No |
ExAC gnomAD |
|
| rs763623314 | 1625 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs763623314 | 1625 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs764828871 | 1626 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs764828871 | 1626 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs764828871 | 1626 | N>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1627 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1483645258 | 1627 | L>V | No |
TOPMed gnomAD |
|
| rs368512848 | 1628 | A>T | No |
ESP ExAC gnomAD |
|
| rs759976196 | 1628 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs777195789 | 1629 | R>C | No |
ExAC gnomAD |
|
| rs771256532 | 1629 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs771256532 | 1629 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs747297506 | 1630 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1566837953 | 1631 | P>R | No | Ensembl | |
| rs773328858 | 1632 | P>L | No |
ExAC gnomAD |
|
| rs772132826 | 1633 | S>T | No |
ExAC gnomAD |
|
|
rs56412851 VAR_040839 |
1633 | S>Y | No |
UniProt 1000Genomes ExAC dbSNP gnomAD |
|
| rs1452095712 | 1634 | R>S | No | TOPMed | |
| rs1360690370 | 1635 | N>K | No | gnomAD | |
| rs562948212 | 1635 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1469993209 | 1637 | P>L | No | gnomAD | |
| rs1157623182 | 1637 | P>S | No |
TOPMed gnomAD |
|
| rs1208737543 | 1640 | S>L | No |
TOPMed gnomAD |
|
| rs1256915996 | 1640 | S>P | No | gnomAD | |
|
rs1287079702 COSM3493934 |
1642 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs757029829 | 1643 | S>A | No |
ExAC gnomAD |
|
| rs1251789116 | 1645 | G>V | No | TOPMed | |
| rs1467901454 | 1646 | G>A | No | gnomAD | |
| rs1467901454 | 1646 | G>E | No | gnomAD | |
| rs80130415 | 1647 | S>L | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1648 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1648 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1891722930 | 1649 | P>A | No | Ensembl | |
| rs763037239 | 1650 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs763037239 | 1650 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1891722668 | 1650 | S>I | No | TOPMed | |
| rs201627729 | 1651 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM3493933 rs746810463 |
1652 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| TCGA novel | 1653 | V>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777479076 | 1653 | V>G | No |
ExAC gnomAD |
|
| rs566836182 | 1653 | V>M | No | Ensembl | |
| rs1891721651 | 1654 | P>A | No |
TOPMed gnomAD |
|
| rs1891721547 | 1654 | P>L | No | TOPMed | |
| rs1891721651 | 1654 | P>S | No |
TOPMed gnomAD |
|
| rs1891721453 | 1655 | L>P | No | TOPMed | |
| rs1370756827 | 1656 | R>K | No |
TOPMed gnomAD |
|
| rs1026746039 | 1657 | S>R | No | Ensembl | |
| rs138426317 | 1658 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1450205910 | 1658 | M>T | No | TOPMed | |
| rs138426317 | 1658 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs754471450 | 1659 | S>F | No |
ExAC gnomAD |
|
| rs1314054576 | 1659 | S>T | No |
TOPMed gnomAD |
|
| rs753465444 | 1660 | D>H | No |
ExAC gnomAD |
|
| rs1382376082 | 1661 | P>A | No | TOPMed | |
| rs1382376082 | 1661 | P>T | No | TOPMed | |
| rs2139339348 | 1662 | D>H | No | Ensembl | |
| rs1453956201 | 1667 | K>R | No |
TOPMed gnomAD |
|
| rs756627458 | 1668 | E>* | No |
ExAC gnomAD |
|
| rs750766228 | 1668 | E>V | No |
ExAC gnomAD |
|
| rs1287785304 | 1669 | P>L | No |
TOPMed gnomAD |
|
| rs750844426 | 1669 | P>S | No |
ExAC gnomAD |
|
| rs781727740 | 1670 | D>E | No |
ExAC gnomAD |
|
| rs1891507761 | 1670 | D>N | No | TOPMed | |
| rs867664425 | 1671 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1891507517 | 1671 | S>T | No | TOPMed | |
| rs867664425 | 1671 | S>W | No |
TOPMed gnomAD |
|
| rs1280193003 | 1673 | S>F | No | gnomAD | |
| rs1891506706 | 1674 | T>N | No | Ensembl | |
| rs1595438845 | 1674 | T>P | No | Ensembl | |
| rs764290721 | 1675 | K>E | No |
ExAC gnomAD |
|
|
rs1595438835 COSM1477380 |
1675 | K>N | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs758672847 | 1675 | K>R | No |
ExAC gnomAD |
|
| rs535677589 | 1676 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1891506186 | 1676 | H>R | No | TOPMed | |
| COSM4846197 | 1677 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768331940 | 1678 | T>A | No | Ensembl | |
| COSM953589 | 1678 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1679 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765191399 | 1680 | S>L | No |
ExAC gnomAD |
|
| rs1891505335 | 1681 | N>S | No | TOPMed | |
| rs1595438792 | 1684 | N>T | No | Ensembl | |
|
RCV001771494 rs2139325341 |
1685 | P>L | No |
ClinVar Ensembl dbSNP |
|
| rs1364479548 | 1685 | P>T | No |
TOPMed gnomAD |
|
| rs767627879 | 1687 | G>C | No |
1000Genomes ExAC gnomAD |
|
| rs767627879 | 1687 | G>S | No |
1000Genomes ExAC gnomAD |
|
| rs1051697726 | 1687 | G>V | No | Ensembl | |
| rs1891504148 | 1688 | P>L | No | gnomAD | |
| rs1473965879 | 1689 | P>L | No | gnomAD | |
| rs1891503899 | 1689 | P>S | No | TOPMed | |
| COSM3493932 | 1691 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4846255 rs1485309676 |
1693 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1485309676 | 1693 | S>F | No | gnomAD | |
| rs760303410 | 1695 | H>D | No |
ExAC TOPMed gnomAD |
|
| rs760303410 | 1695 | H>N | No |
ExAC TOPMed gnomAD |
|
| rs775226335 | 1695 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs760303410 | 1695 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1246903938 | 1696 | R>G | No |
TOPMed gnomAD |
|
| rs562551309 | 1696 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs1891501960 | 1699 | L>P | No | gnomAD | |
| rs1233179130 | 1700 | P>A | No | gnomAD | |
| rs1233179130 | 1700 | P>S | No | gnomAD | |
| rs140640502 | 1701 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1701 | L>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs140640502 | 1701 | L>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs758823144 | 1702 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs758823144 | 1702 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1891500630 | 1703 | G>D | No | TOPMed | |
| rs1891500276 | 1704 | L>P | No | Ensembl | |
| rs1474790652 | 1704 | L>V | No |
TOPMed gnomAD |
|
| rs1891500165 | 1706 | Q>* | No | TOPMed | |
| rs765510912 | 1707 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs765510912 | 1707 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs752816236 | 1707 | P>S | No |
ExAC gnomAD |
|
| rs1459767285 | 1709 | C>G | No | gnomAD | |
| rs1595438671 | 1711 | T>P | No | Ensembl |
1 associated diseases with Q9Y5S2
[MIM: 619841]: Chilton-Okur-Chung neurodevelopmental syndrome (CHOCNS)
A disorder characterized by developmental delay, intellectual disability, hypotonia, and structural brain abnormalities including cerebellar vermis hypoplasia and agenesis or hypoplasia of the corpus callosum. Most patients have behavioral abnormalities, including autism spectrum disorder, attention deficit and hyperactivity disorder, and aggression. About half of patients have dysmorphic facial features. Rare involvement of other organ systems may be present. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A disorder characterized by developmental delay, intellectual disability, hypotonia, and structural brain abnormalities including cerebellar vermis hypoplasia and agenesis or hypoplasia of the corpus callosum. Most patients have behavioral abnormalities, including autism spectrum disorder, attention deficit and hyperactivity disorder, and aggression. About half of patients have dysmorphic facial features. Rare involvement of other organ systems may be present. . Note=The disease is caused by variants affecting the gene represented in this entry.
15 regional properties for Q9Y5S2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | CRIB domain | 1583 - 1618 | IPR000095 |
| domain | Protein kinase domain | 76 - 342 | IPR000719 |
| domain | AGC-kinase, C-terminal | 343 - 413 | IPR000961 |
| domain | Citron homology (CNH) domain | 1240 - 1519 | IPR001180 |
| domain | Pleckstrin homology domain | 1095 - 1216 | IPR001849 |
| domain | Protein kinase C-like, phorbol ester/diacylglycerol-binding domain | 1025 - 1076 | IPR002219 |
| active_site | Serine/threonine-protein kinase, active site | 196 - 208 | IPR008271 |
| domain | Myotonic dystrophy protein kinase, coiled coil | 878 - 939 | IPR014930 |
| binding_site | Protein kinase, ATP binding site | 82 - 105 | IPR017441 |
| domain | Diacylglycerol/phorbol-ester binding | 1023 - 1037 | IPR020454-1 |
| domain | Diacylglycerol/phorbol-ester binding | 1039 - 1048 | IPR020454-2 |
| domain | Diacylglycerol/phorbol-ester binding | 1052 - 1063 | IPR020454-3 |
| domain | Diacylglycerol/phorbol-ester binding | 1064 - 1076 | IPR020454-4 |
| domain | KELK-motif containing domain | 528 - 606 | IPR031597 |
| domain | Serine/threonine-protein kinase MRCK beta, catalytic domain | 3 - 411 | IPR042718 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| actomyosin | Any complex of actin, myosin, and accessory proteins. |
| cell leading edge | The area of a motile cell closest to the direction of movement. |
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
| protein-containing complex binding | Binding to a macromolecular complex. |
| small GTPase binding | Binding to a small monomeric GTPase. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| actomyosin structure organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures containing both actin and myosin or paramyosin. The myosin may be organized into filaments. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. |
| cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures. |
| establishment or maintenance of cell polarity | Any cellular process that results in the specification, formation or maintenance of anisotropic intracellular organization or cell growth patterns. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q28021 | ROCK2 | Rho-associated protein kinase 2 | Bos taurus (Bovine) | SS |
| Q9W1B0 | gek | Serine/threonine-protein kinase Genghis Khan | Drosophila melanogaster (Fruit fly) | SS |
| O75116 | ROCK2 | Rho-associated protein kinase 2 | Homo sapiens (Human) | SS |
| Q13464 | ROCK1 | Rho-associated protein kinase 1 | Homo sapiens (Human) | SS |
| Q5VT25 | CDC42BPA | Serine/threonine-protein kinase MRCK alpha | Homo sapiens (Human) | EV |
| P70336 | Rock2 | Rho-associated protein kinase 2 | Mus musculus (Mouse) | SS |
| Q3UU96 | Cdc42bpa | Serine/threonine-protein kinase MRCK alpha | Mus musculus (Mouse) | SS |
| P70335 | Rock1 | Rho-associated protein kinase 1 | Mus musculus (Mouse) | SS |
| Q80UW5 | Cdc42bpg | Serine/threonine-protein kinase MRCK gamma | Mus musculus (Mouse) | PR |
| Q7TT50 | Cdc42bpb | Serine/threonine-protein kinase MRCK beta | Mus musculus (Mouse) | SS |
| M3TYT0 | ROCK2 | Rho-associated protein kinase 2 | Sus scrofa (Pig) | SS |
| O54874 | Cdc42bpa | Serine/threonine-protein kinase MRCK alpha | Rattus norvegicus (Rat) | SS |
| Q62868 | Rock2 | Rho-associated protein kinase 2 | Rattus norvegicus (Rat) | EV |
| Q63644 | Rock1 | Rho-associated protein kinase 1 | Rattus norvegicus (Rat) | SS |
| Q7TT49 | Cdc42bpb | Serine/threonine-protein kinase MRCK beta | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSAKVRLKKL | EQLLLDGPWR | NESALSVETL | LDVLVCLYTE | CSHSALRRDK | YVAEFLEWAK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PFTQLVKEMQ | LHREDFEIIK | VIGRGAFGEV | AVVKMKNTER | IYAMKILNKW | EMLKRAETAC |
| 130 | 140 | 150 | 160 | 170 | 180 |
| FREERDVLVN | GDCQWITALH | YAFQDENHLY | LVMDYYVGGD | LLTLLSKFED | KLPEDMARFY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IGEMVLAIDS | IHQLHYVHRD | IKPDNVLLDV | NGHIRLADFG | SCLKMNDDGT | VQSSVAVGTP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DYISPEILQA | MEDGMGKYGP | ECDWWSLGVC | MYEMLYGETP | FYAESLVETY | GKIMNHEERF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| QFPSHVTDVS | EEAKDLIQRL | ICSRERRLGQ | NGIEDFKKHA | FFEGLNWENI | RNLEAPYIPD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VSSPSDTSNF | DVDDDVLRNT | EILPPGSHTG | FSGLHLPFIG | FTFTTESCFS | DRGSLKSIMQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SNTLTKDEDV | QRDLEHSLQM | EAYERRIRRL | EQEKLELSRK | LQESTQTVQS | LHGSSRALSN |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SNRDKEIKKL | NEEIERLKNK | IADSNRLERQ | LEDTVALRQE | REDSTQRLRG | LEKQHRVVRQ |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EKEELHKQLV | EASERLKSQA | KELKDAHQQR | KLALQEFSEL | NERMAELRAQ | KQKVSRQLRD |
| 610 | 620 | 630 | 640 | 650 | 660 |
| KEEEMEVATQ | KVDAMRQEMR | RAEKLRKELE | AQLDDAVAEA | SKERKLREHS | ENFCKQMESE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LEALKVKQGG | RGAGATLEHQ | QEISKIKSEL | EKKVLFYEEE | LVRREASHVL | EVKNVKKEVH |
| 730 | 740 | 750 | 760 | 770 | 780 |
| DSESHQLALQ | KEILMLKDKL | EKSKRERHNE | MEEAVGTIKD | KYERERAMLF | DENKKLTAEN |
| 790 | 800 | 810 | 820 | 830 | 840 |
| EKLCSFVDKL | TAQNRQLEDE | LQDLAAKKES | VAHWEAQIAE | IIQWVSDEKD | ARGYLQALAS |
| 850 | 860 | 870 | 880 | 890 | 900 |
| KMTEELEALR | SSSLGSRTLD | PLWKVRRSQK | LDMSARLELQ | SALEAEIRAK | QLVQEELRKV |
| 910 | 920 | 930 | 940 | 950 | 960 |
| KDANLTLESK | LKDSEAKNRE | LLEEMEILKK | KMEEKFRADT | GLKLPDFQDS | IFEYFNTAPL |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| AHDLTFRTSS | ASEQETQAPK | PEASPSMSVA | ASEQQEDMAR | PPQRPSAVPL | PTTQALALAG |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| PKPKAHQFSI | KSFSSPTQCS | HCTSLMVGLI | RQGYACEVCS | FACHVSCKDG | APQVCPIPPE |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| QSKRPLGVDV | QRGIGTAYKG | HVKVPKPTGV | KKGWQRAYAV | VCDCKLFLYD | LPEGKSTQPG |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| VIASQVLDLR | DDEFSVSSVL | ASDVIHATRR | DIPCIFRVTA | SLLGAPSKTS | SLLILTENEN |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| EKRKWVGILE | GLQSILHKNR | LRNQVVHVPL | EAYDSSLPLI | KAILTAAIVD | ADRIAVGLEE |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| GLYVIEVTRD | VIVRAADCKK | VHQIELAPRE | KIVILLCGRN | HHVHLYPWSS | LDGAEGSFDI |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| KLPETKGCQL | MATATLKRNS | GTCLFVAVKR | LILCYEIQRT | KPFHRKFNEI | VAPGSVQCLA |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| VLRDRLCVGY | PSGFCLLSIQ | GDGQPLNLVN | PNDPSLAFLS | QQSFDALCAV | ELESEEYLLC |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| FSHMGLYVDP | QGRRARAQEL | MWPAAPVACS | CSPTHVTVYS | EYGVDVFDVR | TMEWVQTIGL |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| RRIRPLNSEG | TLNLLNCEPP | RLIYFKSKFS | GAVLNVPDTS | DNSKKQMLRT | RSKRRFVFKV |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| PEEERLQQRR | EMLRDPELRS | KMISNPTNFN | HVAHMGPGDG | MQVLMDLPLS | AVPPSQEERP |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| GPAPTNLARQ | PPSRNKPYIS | WPSSGGSEPS | VTVPLRSMSD | PDQDFDKEPD | SDSTKHSTPS |
| 1690 | 1700 | 1710 | |||
| NSSNPSGPPS | PNSPHRSQLP | LEGLEQPACD | T |