Q9Y4X5
Gene name |
ARIH1 |
Protein name |
E3 ubiquitin-protein ligase ARIH1 |
Names |
H7-AP2, HHARI, Monocyte protein 6, MOP-6, Protein ariadne-1 homolog, ARI-1, UbcH7-binding protein, UbcM4-interacting protein, Ubiquitin-conjugating enzyme E2-binding protein 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:25820 |
EC number |
2.3.2.31: Aminoacyltransferases |
Protein Class |
RBR FAMILY RING FINGER AND IBR DOMAIN-CONTAINING (PTHR11685) |
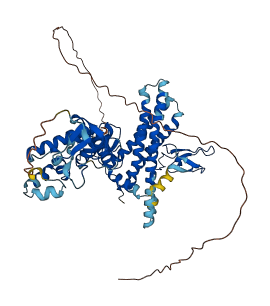
Descriptions
E3 ubiquitin-protein ligase ARIH1 is an E3 ubiquitin-protein ligase, which catalyzes ubiquitination of target proteins together with ubiquitin-conjugating enzyme E2 UBE2L3, and acts as an atypical E3 ubiquitin-protein ligase by working together with cullin-RING ubiquitin ligase (CRL) complexes and initiating ubiquitination of CRL substrates. ARIH1 is autoinhibited by the ariadne domain, which masks the second RING-type zinc finger that contains the active site and inhibits the E3 activity. The inhibition is relieved upon binding to neddylated cullin-RING ubiquitin ligase complexes, which activate the E3 ligase activity of ARIH1.
Autoinhibitory domains (AIDs)
Target domain |
276-347 (RING2 domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
References
- Duda DM et al. (2013) "Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism", Structure (London, England : 1993), 21, 1030-41
- Kelsall IR et al. (2013) "TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes", The EMBO journal, 32, 2848-60
Autoinhibited structure
Activated structure

11 structures for Q9Y4X5
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1WD2 | NMR | - | A | 336-394 | PDB |
| 2M9Y | NMR | - | A | 325-396 | PDB |
| 4KBL | X-ray | 330 A | A/B | 1-557 | PDB |
| 4KC9 | X-ray | 360 A | A | 1-557 | PDB |
| 5TTE | X-ray | 350 A | B | 1-557 | PDB |
| 5UDH | X-ray | 324 A | A/B | 90-557 | PDB |
| 7B5L | EM | 380 A | H | 1-557 | PDB |
| 7B5M | EM | 391 A | H | 1-557 | PDB |
| 7B5N | EM | 360 A | H | 1-557 | PDB |
| 7B5S | EM | 360 A | H | 1-557 | PDB |
| AF-Q9Y4X5-F1 | Predicted | AlphaFoldDB |
167 variants for Q9Y4X5
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
VAR_082646 CA273260089 rs1052050835 RCV001291053 |
15 | E>Q | probable disease-associated variant found in patient with an acute aortic dissection and ascending aortic aneurysm Aortic aneurysm [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt TOPMed dbSNP |
|
rs2063786716 RCV001291431 |
44 | E>G | Aortic aneurysm [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001291430 rs746505361 CA7645516 |
171 | R>* | Aortic aneurysm [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
rs751344394 CA7645402 |
7 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393235936 rs1302485230 |
7 | Y>H | No |
ClinGen TOPMed |
|
|
rs1361234355 CA393235990 |
8 | N>H | No |
ClinGen TOPMed |
|
|
rs781240234 CA7645404 |
8 | N>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1477796553 CA393236038 |
9 | Y>C | No |
ClinGen TOPMed |
|
|
CA393236051 rs1479803832 |
10 | E>Q | No |
ClinGen gnomAD |
|
|
rs1195230372 CA393236099 |
11 | F>L | No |
ClinGen TOPMed |
|
|
rs769816706 CA7645406 |
12 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1428522519 CA393236106 |
12 | D>N | No |
ClinGen TOPMed |
|
|
rs779981165 CA7645407 |
15 | E>A | No |
ClinGen ExAC gnomAD |
|
|
rs779981165 CA393236205 |
15 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs890255198 CA273260090 |
18 | S>C | No |
ClinGen gnomAD |
|
|
rs890255198 CA393236268 |
18 | S>G | No |
ClinGen gnomAD |
|
|
rs1321793234 CA393236299 |
19 | E>K | No |
ClinGen gnomAD |
|
|
rs868117311 CA273260091 |
21 | D>E | No |
ClinGen Ensembl |
|
|
CA7645411 rs768692029 |
22 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs772456417 CA7645415 |
25 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1327083729 CA393236373 |
25 | E>Q | No |
ClinGen TOPMed |
|
|
CA7645416 rs773605993 |
28 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1348781230 CA393236418 |
28 | E>G | No |
ClinGen gnomAD |
|
|
CA7645418 rs761158526 |
29 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA7645417 rs761158526 |
29 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA7645419 rs752084483 |
30 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1490087109 CA393236491 |
32 | D>E | No |
ClinGen gnomAD |
|
|
CA393236505 rs1182181311 |
33 | D>E | No |
ClinGen gnomAD |
|
|
CA393236554 rs1253333685 |
36 | D>E | No |
ClinGen gnomAD |
|
|
CA7645421 rs763608232 |
37 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs751293008 CA7645423 |
39 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA7645422 rs751293008 |
39 | T>N | No |
ClinGen ExAC gnomAD |
|
|
rs946176693 CA273260094 |
41 | D>N | No |
ClinGen TOPMed |
|
|
rs994449992 CA273260095 |
45 | V>A | No |
ClinGen gnomAD |
|
|
CA7645427 rs780042095 |
47 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1430641905 CA393236752 |
49 | E>* | No |
ClinGen gnomAD |
|
|
CA7645428 rs749268378 |
49 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA393236754 rs1595845215 |
49 | E>G | No |
ClinGen Ensembl |
|
|
rs768801971 CA7645429 |
50 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA273260096 rs1042181947 |
51 | G>W | No |
ClinGen TOPMed |
|
|
rs1312645720 CA393236805 |
53 | G>S | No |
ClinGen gnomAD |
|
|
rs1358544322 CA393236815 |
54 | V>I | No |
ClinGen gnomAD |
|
|
CA393236833 rs1239114198 |
55 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
CA393236840 rs1184730148 |
56 | G>R | No |
ClinGen TOPMed |
|
|
rs1000583730 CA273260097 |
57 | E>Q | No |
ClinGen TOPMed |
|
|
CA7645433 rs773561251 |
58 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1177781521 CA393236885 |
59 | D>N | No |
ClinGen TOPMed |
|
|
CA393236943 rs1426082091 |
64 | G>R | No |
ClinGen gnomAD |
|
|
rs777220733 CA7645436 |
66 | T>M | No |
ClinGen ExAC gnomAD |
|
|
CA393236976 rs1450537917 |
69 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1013967251 CA273260099 |
71 | G>S | No |
ClinGen Ensembl |
|
|
CA393236992 rs1264967111 |
72 | S>C | No |
ClinGen TOPMed |
|
|
CA393236999 rs774057607 |
73 | A>P | No |
ClinGen ExAC gnomAD |
|
|
CA7645439 rs774057607 |
73 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1303516615 CA393237004 |
74 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA7645442 rs559502830 |
76 | P>L | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs767341707 CA7645441 |
76 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1228917143 CA393237018 |
77 | G>S | No |
ClinGen gnomAD |
|
|
rs755947974 CA7645446 |
78 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA393237049 rs1199225308 |
82 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA393237048 rs1199225308 |
82 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA393237056 rs1465600303 |
83 | G>C | No |
ClinGen gnomAD |
|
|
rs12902861 CA273260104 |
84 | G>C | No |
ClinGen Ensembl |
|
|
rs753843976 CA7645456 |
85 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA273260105 rs967532979 |
86 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1417513681 CA393237078 |
87 | G>D | No |
ClinGen TOPMed |
|
|
rs1297238945 CA393237076 |
87 | G>R | No |
ClinGen TOPMed |
|
|
CA273260106 rs977706882 |
88 | G>S | No |
ClinGen Ensembl |
|
|
rs1379429183 CA393237087 |
89 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
RCV002056684 RCV000455671 rs375614248 |
90 | G>missing | No |
ClinVar dbSNP |
|
|
CA393237100 rs1375923364 |
91 | P>S | No |
ClinGen TOPMed |
|
|
rs1464648760 CA393237106 |
92 | G>W | No |
ClinGen gnomAD |
|
|
rs1405301183 CA393237114 |
93 | H>R | No |
ClinGen gnomAD |
|
|
rs1481483194 CA393237120 |
94 | E>Q | No |
ClinGen TOPMed |
|
|
CA273260108 rs957384868 |
95 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1359687603 CA393237167 |
100 | R>C | No |
ClinGen gnomAD |
|
|
CA7645461 rs368784844 |
100 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1206162417 CA393237174 |
101 | Y>C | No |
ClinGen TOPMed |
|
|
rs921251553 CA273260109 |
102 | E>K | No |
ClinGen gnomAD |
|
|
CA273260110 rs12903022 |
107 | E>D | No |
ClinGen Ensembl |
|
|
rs1210904578 CA393237232 |
110 | L>V | No |
ClinGen gnomAD |
|
|
CA393237253 rs1191965773 |
113 | M>L | No |
ClinGen gnomAD |
|
|
CA273260112 rs867712471 |
118 | R>L | No |
ClinGen Ensembl |
|
|
CA7645467 rs746369007 |
121 | N>K | No |
ClinGen ExAC gnomAD |
|
|
CA7645469 rs770477644 |
122 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771736875 CA7645492 |
130 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393242616 rs771736875 |
130 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393242655 rs1297406432 |
136 | S>G | No |
ClinGen TOPMed |
|
|
rs147408829 CA273265289 |
139 | N>S | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1243956076 CA393235448 |
154 | L>V | No |
ClinGen gnomAD |
|
|
rs933413352 CA273268323 |
162 | H>R | No |
ClinGen TOPMed |
|
|
rs769334669 CA7645515 |
167 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769334669 CA7645514 |
167 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7645517 rs770559164 |
171 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs148941976 CA7645518 |
173 | R>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs969696237 CA273268324 |
173 | R>H | No |
ClinGen gnomAD |
|
|
CA393235841 rs979829613 |
174 | Q>H | No |
ClinGen Ensembl |
|
|
rs1251423210 CA393235886 |
176 | N>S | No |
ClinGen TOPMed |
|
|
CA7645519 rs759288383 |
177 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA7645521 rs775316269 |
180 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7645523 rs764009252 |
183 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1000868639 CA273268326 |
191 | L>V | No |
ClinGen TOPMed |
|
|
CA273268327 rs779405166 |
192 | N>K | No |
ClinGen TOPMed |
|
|
CA393236276 rs1345775312 |
196 | S>L | No |
ClinGen gnomAD |
|
|
CA7645537 rs769496771 |
197 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs1295878502 CA393237641 |
198 | F>L | No |
ClinGen gnomAD |
|
|
CA393237676 rs1345038178 |
202 | E>Q | No |
ClinGen gnomAD |
|
|
CA273269551 rs961904194 |
209 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
rs961904194 CA273269550 |
209 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
CA393237876 rs1228878159 |
213 | S>N | No |
ClinGen gnomAD |
|
|
rs1286785152 CA393238218 |
229 | I>T | No |
ClinGen TOPMed |
|
|
CA393238283 rs1343881622 |
234 | H>N | No |
ClinGen TOPMed |
|
|
CA7645560 rs748015574 |
235 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA393238472 rs1181805525 |
246 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
CA7645576 rs754711191 |
247 | R>C | No |
ClinGen ExAC TOPMed |
|
|
CA7645577 rs778561142 |
247 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs147551270 CA7645578 |
251 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1392004426 CA393239750 |
251 | D>N | No |
ClinGen TOPMed |
|
|
rs1461577237 CA393239907 |
262 | I>V | No |
ClinGen gnomAD |
|
|
CA393240186 rs1405057056 |
270 | N>S | No |
ClinGen gnomAD |
|
|
rs1454157851 CA393240191 |
271 | R>Q | No |
ClinGen gnomAD |
|
|
rs778633164 CA7645597 |
272 | L>R | No |
ClinGen ExAC gnomAD |
|
|
rs149030231 CA273270415 |
280 | D>G | No |
ClinGen ESP |
|
|
rs1218416217 CA393240249 |
280 | D>H | No |
ClinGen TOPMed |
|
|
rs758299803 CA7645599 |
283 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs777749802 CA7645600 |
288 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA393240354 rs1271592185 |
288 | Q>R | No |
ClinGen TOPMed |
|
|
CA393240410 rs1339411218 |
292 | A>T | No |
ClinGen TOPMed |
|
|
rs781125737 CA7645603 |
294 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA393240576 rs1332093999 |
294 | P>L | No |
ClinGen TOPMed |
|
|
rs1259128502 COSM1374415 CA393240585 |
296 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1259128502 CA393240584 |
296 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs769846251 CA7645605 |
296 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769846251 CA7645606 |
296 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393240607 rs1463886403 |
299 | C>S | No |
ClinGen TOPMed |
|
|
rs1417583080 CA393240617 |
301 | R>C | No |
ClinGen TOPMed |
|
|
rs1188543265 CA393240620 |
301 | R>H | No |
ClinGen TOPMed |
|
|
rs1320295436 CA393240801 |
321 | K>N | No |
ClinGen TOPMed |
|
|
CA7645640 rs757137026 |
326 | K>T | No |
ClinGen ExAC |
|
|
CA393240891 rs1595871915 |
333 | T>P | No |
ClinGen Ensembl |
|
|
rs750348642 CA7645642 |
335 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs192253156 CA273271038 |
357 | C>Y | No |
ClinGen 1000Genomes |
|
|
rs969550607 CA273271039 |
365 | Q>* | No |
ClinGen Ensembl |
|
|
rs573902027 CA7645661 |
370 | E>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1284449604 CA393241194 |
373 | W>R | No |
ClinGen gnomAD |
|
|
CA7645686 rs753804913 |
391 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA7645687 rs759517486 |
399 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA7645734 rs762126728 |
406 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA393241459 rs762126728 |
406 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA7645735 rs767859277 |
409 | A>E | No |
ClinGen ExAC gnomAD |
|
|
rs753701203 CA7645738 |
420 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs777142325 CA7645741 |
443 | E>G | No |
ClinGen ExAC |
|
|
CA273272166 rs113869196 |
459 | K>R | No |
ClinGen Ensembl |
|
|
CA7645747 rs779944062 |
483 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs768710024 CA7645749 |
484 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1203862513 CA393242169 |
503 | E>K | No |
ClinGen gnomAD |
|
|
CA7645776 rs753167225 |
521 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1447655771 CA393242349 |
528 | K>M | No |
ClinGen gnomAD |
|
|
COSM382806 CA393242415 rs1276382860 |
535 | R>L | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs770168604 CA7645801 |
538 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs1370979485 CA393242436 |
539 | L>S | No |
ClinGen TOPMed |
|
|
rs1218251335 CA393242453 |
541 | Q>H | No |
ClinGen gnomAD |
|
|
rs1213846918 CA393242474 |
544 | H>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1213846918 CA393242472 |
544 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
CA393242495 rs1348294540 |
547 | Y>C | No |
ClinGen TOPMed |
|
|
rs1245309692 CA393242551 |
554 | Y>* | No |
ClinGen gnomAD |
|
|
CA393242570 rs1477923543 |
557 | D>Y | No |
ClinGen gnomAD |
1 associated diseases with Q9Y4X5
Without disease ID
6 regional properties for Q9Y4X5
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Zinc finger, RING-type | 186 - 234 | IPR001841-1 |
| domain | Zinc finger, RING-type | 276 - 347 | IPR001841-2 |
| domain | IBR domain | 256 - 317 | IPR002867-1 |
| domain | IBR domain | 325 - 389 | IPR002867-2 |
| domain | TRIAD supradomain | 182 - 393 | IPR044066 |
| domain | Ariadne domain | 403 - 524 | IPR045840 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.31 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11685 | RBR FAMILY RING FINGER AND IBR DOMAIN-CONTAINING |
| PANTHER Subfamily | PTHR11685:SF212 | E3 UBIQUITIN-PROTEIN LIGASE ARIH1 |
| PANTHER Protein Class |
ubiquitin-protein ligase
protein modifying enzyme |
|
| PANTHER Pathway Category | No pathway information available | |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| Cajal body | A class of nuclear body, first seen after silver staining by Ramon y Cajal in 1903, enriched in small nuclear ribonucleoproteins, and certain general RNA polymerase II transcription factors; ultrastructurally, they appear as a tangle of coiled, electron-dense threads roughly 0.5 micrometers in diameter; involved in aspects of snRNP biogenesis; the protein coilin serves as a marker for Cajal bodies. Some argue that Cajal bodies are the sites for preassembly of transcriptosomes, unitary particles involved in transcription and processing of RNA. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| Lewy body | Cytoplasmic, spherical inclusion commonly found in damaged neurons, and composed of abnormally phosphorylated, neurofilament proteins aggregated with ubiquitin and alpha-synuclein. |
| nuclear body | Extra-nucleolar nuclear domains usually visualized by confocal microscopy and fluorescent antibodies to specific proteins. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| ubiquitin ligase complex | A protein complex that includes a ubiquitin-protein ligase and enables ubiquitin protein ligase activity. The complex also contains other proteins that may confer substrate specificity on the complex. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ubiquitin conjugating enzyme binding | Binding to a ubiquitin conjugating enzyme, any of the E2 proteins. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
| ubiquitin-like protein transferase activity | Catalysis of the transfer of a ubiquitin-like from one protein to another via the reaction X-ULP + Y --> Y-ULP + X, where both X-ULP and Y-ULP are covalent linkages. ULP represents a ubiquitin-like protein. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
| zinc ion binding | Binding to a zinc ion (Zn). |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of a ubiquitin group, or multiple ubiquitin groups, to the protein. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A2VEA3 | ARIH1 | E3 ubiquitin-protein ligase ARIH1 | Bos taurus (Bovine) | SS |
| Q94981 | ari-1 | E3 ubiquitin-protein ligase ariadne-1 | Drosophila melanogaster (Fruit fly) | SS |
| O76924 | ari-2 | Potential E3 ubiquitin-protein ligase ariadne-2 | Drosophila melanogaster (Fruit fly) | SS |
| O60260 | PRKN | E3 ubiquitin-protein ligase parkin | Homo sapiens (Human) | EV |
| O95376 | ARIH2 | E3 ubiquitin-protein ligase ARIH2 | Homo sapiens (Human) | EV |
| Q9Z1K5 | Arih1 | E3 ubiquitin-protein ligase ARIH1 | Mus musculus (Mouse) | SS |
| Q9Z1K6 | Arih2 | E3 ubiquitin-protein ligase ARIH2 | Mus musculus (Mouse) | SS |
| O01965 | ari-1.1 | E3 ubiquitin-protein ligase ari-1.1 | Caenorhabditis elegans | SS |
| Q22431 | ari-2 | Potential E3 ubiquitin-protein ligase ariadne-2 | Caenorhabditis elegans | SS |
| B1H1E4 | arih1 | E3 ubiquitin-protein ligase arih1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q6NW85 | arih1l | E3 ubiquitin-protein ligase arih1l | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q6PFJ9 | arih1 | E3 ubiquitin-protein ligase arih1 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDSDEGYNYE | FDEDEECSEE | DSGAEEEEDE | DDDEPDDDTL | DLGEVELVEP | GLGVGGERDG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LLCGETGGGG | GSALGPGGGG | GGGGGGGGGG | PGHEQEEDYR | YEVLTAEQIL | QHMVECIREV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NEVIQNPATI | TRILLSHFNW | DKEKLMERYF | DGNLEKLFAE | CHVINPSKKS | RTRQMNTRSS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| AQDMPCQICY | LNYPNSYFTG | LECGHKFCMQ | CWSEYLTTKI | MEEGMGQTIS | CPAHGCDILV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DDNTVMRLIT | DSKVKLKYQH | LITNSFVECN | RLLKWCPAPD | CHHVVKVQYP | DAKPVRCKCG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RQFCFNCGEN | WHDPVKCKWL | KKWIKKCDDD | SETSNWIAAN | TKECPKCHVT | IEKDGGCNHM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VCRNQNCKAE | FCWVCLGPWE | PHGSAWYNCN | RYNEDDAKAA | RDAQERSRAA | LQRYLFYCNR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| YMNHMQSLRF | EHKLYAQVKQ | KMEEMQQHNM | SWIEVQFLKK | AVDVLCQCRA | TLMYTYVFAF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YLKKNNQSII | FENNQADLEN | ATEVLSGYLE | RDISQDSLQD | IKQKVQDKYR | YCESRRRVLL |
| 550 | |||||
| QHVHEGYEKD | LWEYIED |