Q9Y4I1
Gene name |
MYO5A (MYH12) |
Protein name |
Unconventional myosin-Va |
Names |
Dilute myosin heavy chain, non-muscle , Myosin heavy chain 12 , Myosin-12 , Myoxin |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4644 |
EC number |
|
Protein Class |
|
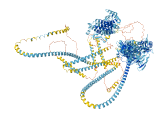
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1446-1830 (Globular tail domain, GTD) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

8 structures for Q9Y4I1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4D07 | X-ray | 185 A | B | 1275-1297 | PDB |
| 4J5L | X-ray | 220 A | A/B | 1448-1855 | PDB |
| 4LLI | X-ray | 220 A | A/B | 1467-1855 | PDB |
| 4LX1 | X-ray | 187 A | A/B | 1464-1855 | PDB |
| 4LX2 | X-ray | 150 A | A | 1464-1855 | PDB |
| 5JCY | X-ray | 180 A | A | 1464-1855 | PDB |
| 5JCZ | X-ray | 206 A | B/C/E | 1464-1855 | PDB |
| AF-Q9Y4I1-F1 | Predicted | AlphaFoldDB |
1530 variants for Q9Y4I1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs2043479059 RCV002248386 |
155 | R>* | Griscelli syndrome type 1 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001779923 rs1724577 RCV002074075 |
362 | E>D | Griscelli syndrome type 1 [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs2042532335 RCV001252333 |
398 | N>K | Intellectual disability [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV002248385 rs2141121140 |
704 | Q>* | Griscelli syndrome type 1 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs764371254 RCV000015113 |
778 | R>* | Griscelli syndrome type 1 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC dbSNP gnomAD |
|
RCV000919256 RCV001333314 rs752461164 |
826 | R>H | Griscelli syndrome type 1 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001252331 rs1252205617 |
827 | M>I | Intellectual disability [ClinVar] | Yes |
ClinVar TOPMed dbSNP |
|
COSM962831 COSM4868402 RCV001809215 rs2141084327 |
1015 | R>* | Variant assessed as Somatic; HIGH impact. Griscelli syndrome type 1 endometrium [NCI-TCGA, ClinVar, Cosmic] | Yes |
NCI-TCGA Cosmic cosmic curated ClinVar Ensembl dbSNP |
|
RCV000723271 rs751411629 |
1033 | L>W | Griscelli syndrome type 1 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001731727 CA7568513 rs56132571 RCV000502933 RCV002527267 RCV001252332 RCV000658717 |
1046 | V>M | Griscelli syndrome type 1 Intellectual disability Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001911804 RCV002555283 rs779402364 |
1087 | R>H | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001355975 RCV002547625 rs199984961 |
1126 | F>C | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000247971 VAR_010645 RCV000678394 RCV001537531 rs1058219 CA7568330 |
1246 | R>C | Griscelli syndrome type 1 [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000490925 rs1114167290 CA392508824 |
1400 | S>R | Dystonic disorder [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000908726 RCV002540781 rs147898420 RCV001252330 |
1453 | P>S | Intellectual disability Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001374539 rs2038687080 |
1506 | P>A | Hereditary breast ovarian cancer syndrome [ClinVar] | Yes |
ClinVar TOPMed dbSNP |
| rs1263769904 | 2 | A>S | No | gnomAD | |
| rs2077772649 | 3 | A>T | No | gnomAD | |
| rs1375361419 | 5 | E>D | No |
TOPMed gnomAD |
|
| rs2077772566 | 5 | E>G | No | Ensembl | |
| rs1344107033 | 6 | L>V | No | gnomAD | |
| rs780931670 | 8 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs780931670 | 8 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs754152277 | 11 | A>S | No |
ExAC gnomAD |
|
| rs1375276866 | 12 | R>G | No |
TOPMed gnomAD |
|
| rs764703163 | 12 | R>S | No |
ExAC gnomAD |
|
| rs1401011577 | 13 | V>I | No |
TOPMed gnomAD |
|
| rs1460851762 | 15 | I>V | No | TOPMed | |
| rs369664207 | 16 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1285512768 | 16 | P>L | No | gnomAD | |
| rs1285512768 | 16 | P>R | No | gnomAD | |
| rs753155018 | 18 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2075580692 | 19 | E>G | No |
TOPMed gnomAD |
|
|
COSM4055588 COSM4055589 |
22 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1437490319 | 24 | S>T | No |
TOPMed gnomAD |
|
| rs1439684979 | 29 | K>Q | No | gnomAD | |
| rs963818562 | 29 | K>R | No | Ensembl | |
| rs2075580186 | 31 | Y>F | No |
TOPMed gnomAD |
|
| rs2075580117 | 32 | K>R | No | TOPMed | |
| rs1371147294 | 35 | D>N | No | gnomAD | |
| rs774614542 | 36 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs2075579875 | 37 | V>F | No |
TOPMed gnomAD |
|
| rs2075579818 | 38 | L>F | No | gnomAD | |
| rs2075579695 | 42 | L>F | No | gnomAD | |
| rs1159660293 | 43 | E>K | No |
TOPMed gnomAD |
|
| rs776264175 | 46 | K>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 46 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1595664425 | 47 | D>V* | No | Ensembl | |
| rs955401113 | 48 | L>S | No | Ensembl | |
| rs2141290476 | 48 | L>V | No | Ensembl | |
| rs1312021115 | 49 | E>K | No | gnomAD | |
| rs778023554 | 50 | Y>C | No |
ExAC gnomAD |
|
| rs2075447314 | 51 | H>R | No | TOPMed | |
| rs1029639593 | 51 | H>Y | No |
TOPMed gnomAD |
|
| rs753215872 | 53 | D>H | No |
ExAC gnomAD |
|
| rs767997640 | 55 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs766191267 | 55 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs751606879 | 61 | H>Y | No |
ExAC TOPMed gnomAD |
|
|
COSM270851 rs1330819225 |
63 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
|
rs766669585 COSM3690437 COSM3690438 |
63 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs763101944 | 67 | I>L | No |
ExAC gnomAD |
|
| rs763101944 | 67 | I>V | No |
ExAC gnomAD |
|
|
COSM6077727 COSM6077728 |
73 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1254997382 | 75 | T>I | No | TOPMed | |
| rs760491276 | 79 | Y>C | No |
ExAC gnomAD |
|
| rs1306485767 | 82 | E>Q | No | gnomAD | |
| rs1295641141 | 83 | P>A | No | gnomAD | |
| rs775628675 | 84 | A>S | No |
ExAC gnomAD |
|
| rs1292553032 | 87 | H>R | No |
TOPMed gnomAD |
|
| rs1275735156 | 89 | L>I | No | gnomAD | |
| rs759161061 | 90 | R>K | No |
ExAC gnomAD |
|
| rs1258207136 | 91 | V>I | No |
TOPMed gnomAD |
|
|
rs2075445471 COSM962856 COSM4252132 |
92 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs199674947 | 92 | R>H | No |
1000Genomes ExAC gnomAD |
|
| rs1461665786 | 94 | I>V | No | gnomAD | |
|
COSM4898198 COSM4898199 |
96 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs748927050 | 97 | K>R | No |
ExAC TOPMed gnomAD |
|
|
COSM3969158 COSM3969159 |
98 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1234793085 | 99 | I>T | No | gnomAD | |
| rs541009526 | 101 | T>M | No |
1000Genomes gnomAD |
|
| rs762509740 | 105 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs769456233 | 108 | V>A | No | ExAC | |
|
COSM4947483 COSM75535 rs1487252374 |
109 | A>G | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs2075385494 | 109 | A>T | No | Ensembl | |
| rs748560806 | 110 | I>T | No |
ExAC gnomAD |
|
| rs1280213902 | 110 | I>V | No | TOPMed | |
|
COSM4867444 COSM962855 |
111 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2075385158 | 111 | N>S | No | gnomAD | |
| rs760727634 | 113 | Y>C | No | TOPMed | |
| rs2075384979 | 114 | E>K | No | Ensembl | |
| rs768965052 | 115 | Q>* | No |
ExAC gnomAD |
|
| rs768965052 | 115 | Q>K | No |
ExAC gnomAD |
|
| TCGA novel | 116 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1336825272 | 116 | L>Q | No | gnomAD | |
| rs747516586 | 118 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1318642737 | 118 | I>V | No | gnomAD | |
| rs780549888 | 122 | D>G | No |
ExAC gnomAD |
|
|
COSM962854 COSM4863769 |
123 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758468634 | 124 | I>T | No |
ExAC gnomAD |
|
| rs750515250 | 126 | A>E | No |
ExAC gnomAD |
|
| rs750515250 | 126 | A>V | No |
ExAC gnomAD |
|
| rs779047860 | 128 | S>N | No |
ExAC gnomAD |
|
| rs2075384292 | 132 | M>V | No | TOPMed | |
| rs2075384235 | 135 | M>L | No | TOPMed | |
| rs1232916707 | 135 | M>R | No |
TOPMed gnomAD |
|
| rs1595659134 | 137 | P>Q | No | Ensembl | |
| rs1443134028 | 139 | I>M | No | gnomAD | |
| rs757535360 | 140 | F>Y | No |
ExAC gnomAD |
|
| rs2075383901 | 143 | A>T | No | TOPMed | |
| rs2075383846 | 143 | A>V | No | TOPMed | |
| rs1454640295 | 144 | E>A | No | gnomAD | |
| rs2075383733 | 147 | Y>S | No | TOPMed | |
|
TCGA novel rs754065070 |
148 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC gnomAD |
| rs1337491186 | 150 | M>T | No | Ensembl | |
| rs2075383572 | 150 | M>V | No | Ensembl | |
| rs2075383461 | 151 | A>V | No | Ensembl | |
| rs2075383408 | 152 | R>K | No | gnomAD | |
| rs776320957 | 153 | D>G | No |
ExAC gnomAD |
|
| rs2043479899 | 153 | D>N | No | gnomAD | |
| rs776320957 | 153 | D>V | No |
ExAC gnomAD |
|
| rs1567105995 | 154 | E>K | No | TOPMed | |
| rs1228854448 | 155 | R>L | No | gnomAD | |
| rs1228854448 | 155 | R>P | No | gnomAD | |
| TCGA novel | 155 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 157 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 158 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs968920076 | 160 | I>F | No | gnomAD | |
| rs968920076 | 160 | I>V | No | gnomAD | |
|
COSM3887142 COSM3887143 |
161 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774327292 | 161 | V>I | No |
ExAC gnomAD |
|
| rs746418214 | 163 | G>R | No |
ExAC gnomAD |
|
| rs1446775765 | 166 | G>E | No |
TOPMed gnomAD |
|
| rs2043477519 | 167 | A>S | No | Ensembl | |
| rs1292140531 | 172 | S>L | No | TOPMed | |
|
COSM5623605 COSM5623604 |
175 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2141252698 | 176 | A>S | No | Ensembl | |
|
COSM962853 rs2043476537 COSM4873728 |
176 | A>V | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed gnomAD |
| rs2043476219 | 177 | M>L | No | TOPMed | |
| rs2043476063 | 177 | M>T | No | TOPMed | |
|
COSM4962574 rs778178180 COSM4962575 |
178 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs2043475692 | 178 | R>Q | No |
TOPMed gnomAD |
|
| rs779929736 | 181 | A>T | No |
ExAC gnomAD |
|
| rs2043475166 | 188 | S>N | No | TOPMed | |
| rs750385472 | 189 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs553305728 | 190 | A>V | No | 1000Genomes | |
| rs1216183008 | 191 | N>S | No | gnomAD | |
| rs1390327109 | 192 | V>L | No | gnomAD | |
|
rs201017976 RCV001940425 |
195 | K>R | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs3208599 | 198 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs1333655292 | 200 | N>S | No |
TOPMed gnomAD |
|
| rs2043473584 | 201 | P>S | No |
TOPMed gnomAD |
|
| rs762982003 | 202 | I>T | No | Ensembl | |
| rs760402593 | 202 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1397990286 | 203 | M>T | No | gnomAD | |
| rs2141235451 | 206 | I>T | No | Ensembl | |
| rs2043199841 | 206 | I>V | No | TOPMed | |
| rs1037859131 | 210 | K>I | No | Ensembl | |
| rs1468919484 | 212 | T>N | No | gnomAD | |
| rs767064853 | 212 | T>S | No | ExAC | |
| rs2043199350 | 213 | R>G | No | TOPMed | |
| rs1255460313 | 215 | D>Y | No |
TOPMed gnomAD |
|
| rs1446850592 | 219 | R>H | No |
TOPMed gnomAD |
|
| rs1446850592 | 219 | R>L | No |
TOPMed gnomAD |
|
| rs1479522952 | 223 | Y>C | No |
TOPMed gnomAD |
|
| rs751949143 | 224 | I>L | No |
ExAC gnomAD |
|
| rs1224038581 | 226 | I>V | No |
TOPMed gnomAD |
|
| rs1355434523 | 229 | D>E | No | gnomAD | |
| rs940861602 | 230 | K>E | No | Ensembl | |
|
COSM4873397 COSM962852 |
231 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2043198207 | 231 | R>K | No | Ensembl | |
| rs2043198084 | 231 | R>S | No | Ensembl | |
| rs2043198207 | 231 | R>T | No | Ensembl | |
|
rs1232932761 COSM5955496 COSM276230 |
233 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs763585223 | 233 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs773788411 | 235 | I>L | No |
ExAC TOPMed gnomAD |
|
| COSM470814 | 236 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs553102801 | 239 | M>I | No |
1000Genomes ExAC gnomAD |
|
| rs933573569 | 240 | R>G | No | TOPMed | |
|
COSM4864675 COSM962851 |
240 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM5604182 COSM5604183 |
243 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs371824735 | 243 | L>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs921934720 | 245 | E>Q | No | Ensembl | |
| rs776975716 | 247 | S>C | No |
ExAC gnomAD |
|
| rs2043196186 | 251 | F>L | No | Ensembl | |
| rs2043196047 | 252 | Q>* | No | Ensembl | |
| TCGA novel | 252 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs969696190 | 255 | E>D | No | Ensembl | |
| rs2141227282 | 256 | E>G | No | Ensembl | |
| rs2043089606 | 257 | R>G | No | TOPMed | |
| TCGA novel | 258 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 259 | Y>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 262 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760921577 | 263 | Y>C | No |
ExAC gnomAD |
|
| rs1257357255 | 265 | L>I | No | gnomAD | |
|
TCGA novel rs2043089041 |
266 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
|
TCGA novel rs2043088906 |
267 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| TCGA novel | 267 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749069988 | 268 | S>L | No |
ExAC gnomAD |
|
| rs2043088494 | 269 | A>T | No | TOPMed | |
| rs773268848 | 272 | P>S | No |
ExAC gnomAD |
|
| rs367598685 | 276 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs780596707 | 276 | M>T | No |
ExAC gnomAD |
|
| rs367598685 | 276 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1415114909 | 278 | R>* | No |
TOPMed gnomAD |
|
| rs754471451 | 278 | R>L | No |
ExAC TOPMed gnomAD |
|
|
COSM4875123 COSM962850 |
278 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs754471451 COSM3355978 COSM962849 |
278 | R>Q | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2043087600 | 280 | G>R | No | Ensembl | |
| rs1323226475 | 281 | N>H | No | gnomAD | |
| rs930212364 | 282 | A>T | No |
TOPMed gnomAD |
|
| rs1391033624 | 282 | A>V | No | gnomAD | |
| rs772903184 | 284 | N>D | No |
ExAC TOPMed gnomAD |
|
|
CA209719 RCV000195114 rs370600193 RCV002517102 |
284 | N>K | No |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
TCGA novel rs2043053306 |
288 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1438756318 | 288 | T>I | No | gnomAD | |
| rs368056746 | 290 | Q>P | No |
ESP ExAC gnomAD |
|
| rs368056746 | 290 | Q>R | No |
ESP ExAC gnomAD |
|
| TCGA novel | 291 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2043052655 | 292 | G>V | No | Ensembl | |
| rs746511998 | 293 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1156539774 | 294 | P>A | No |
TOPMed gnomAD |
|
| rs1168413741 | 296 | I>T | No |
1000Genomes TOPMed |
|
| rs2141224678 | 297 | E>Q | No | Ensembl | |
| rs1418123932 | 297 | E>V | No |
TOPMed gnomAD |
|
| rs1180002089 | 298 | G>A | No |
TOPMed gnomAD |
|
| rs1180002089 | 298 | G>E | No |
TOPMed gnomAD |
|
| rs779892139 | 298 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1162667312 | 299 | V>L | No |
TOPMed gnomAD |
|
| rs1466696738 | 300 | D>A | No |
TOPMed gnomAD |
|
| rs1466696738 | 300 | D>G | No |
TOPMed gnomAD |
|
| rs1466696738 | 300 | D>V | No |
TOPMed gnomAD |
|
| rs977169640 | 301 | D>G | No | TOPMed | |
| rs977169640 | 301 | D>V | No | TOPMed | |
| rs1245689676 | 301 | D>Y | No | gnomAD | |
| rs1252309804 | 302 | A>T | No | gnomAD | |
| rs1406941707 | 302 | A>V | No | TOPMed | |
| rs2043050391 | 304 | E>V | No | TOPMed | |
| rs1196110592 | 305 | M>L | No |
TOPMed gnomAD |
|
| rs991648930 | 306 | A>T | No |
TOPMed gnomAD |
|
| rs771826580 | 306 | A>V | No | ExAC | |
| rs1258716693 | 307 | H>Y | No |
TOPMed gnomAD |
|
| rs1308541954 | 308 | T>A | No |
TOPMed gnomAD |
|
| rs112325631 | 309 | R>S | No |
TOPMed gnomAD |
|
| rs2141224487 | 311 | A>V | No | Ensembl | |
| rs1342756750 | 317 | I>V | No | gnomAD | |
| rs2042959785 | 319 | E>A | No | TOPMed | |
| rs1432445031 | 321 | H>Y | No | gnomAD | |
| rs778295134 | 322 | Q>* | No |
ExAC gnomAD |
|
|
RCV001767782 rs1172688121 |
323 | M>T | No |
ClinVar TOPMed dbSNP gnomAD |
|
|
COSM4897702 COSM4897701 |
324 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756593443 | 324 | G>E | No |
ExAC gnomAD |
|
|
COSM4705001 COSM4705002 |
325 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM1373515 COSM4822530 |
327 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs548669169 | 327 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs781250248 | 328 | I>T | No |
ExAC gnomAD |
|
| rs755024855 | 329 | L>F | No |
ExAC gnomAD |
|
| TCGA novel | 329 | L>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1265026489 | 330 | A>G | No |
TOPMed gnomAD |
|
| rs1265026489 | 330 | A>V | No |
TOPMed gnomAD |
|
| rs751783560 | 331 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs766618790 | 332 | I>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 333 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 334 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753662729 | 334 | H>Y | No |
ExAC gnomAD |
|
| rs1251298304 | 335 | L>F | No |
TOPMed gnomAD |
|
| rs2042957636 | 337 | N>S | No | gnomAD | |
| rs775503661 | 338 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1280306972 | 338 | V>I | No | TOPMed | |
| rs2042957188 | 342 | S>P | No | TOPMed | |
| rs1414508015 | 342 | S>Y | No | gnomAD | |
| rs2042956980 | 343 | R>* | No | Ensembl | |
| rs527359862 | 343 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1182793522 | 345 | A>V | No |
TOPMed gnomAD |
|
|
COSM3690644 COSM3690645 |
346 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770589752 | 349 | T>A | No |
ExAC gnomAD |
|
| rs995381916 | 350 | I>L | No |
TOPMed gnomAD |
|
| rs995381916 | 350 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs373522085 | 351 | P>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1425621215 | 351 | P>S | No | gnomAD | |
| rs765760929 | 352 | P>L | No |
ExAC gnomAD |
|
| rs765760929 | 352 | P>R | No |
ExAC gnomAD |
|
| rs1251743754 | 353 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2042540001 | 353 | K>N | No | Ensembl | |
| rs374157207 | 353 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs772790906 | 354 | H>Y | No |
ExAC gnomAD |
|
| rs770198139 | 356 | P>L | No |
ExAC gnomAD |
|
| TCGA novel | 357 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2042539417 | 357 | L>V | No | TOPMed | |
| rs748503608 | 358 | C>G | No |
ExAC gnomAD |
|
| rs777162187 | 359 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs769184092 | 362 | E>G | No |
ExAC gnomAD |
|
| rs769184092 | 362 | E>V | No |
ExAC gnomAD |
|
| rs2042538397 | 364 | M>L | No | TOPMed | |
|
rs758345429 RCV000192795 CA205865 |
365 | G>D | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs779265368 | 366 | V>A | No |
ExAC gnomAD |
|
| rs746108435 | 366 | V>M | No |
ExAC gnomAD |
|
| rs563513014 | 368 | Y>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2042537404 | 368 | Y>H | No | Ensembl | |
| rs563513014 | 368 | Y>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767407902 | 369 | E>V | No |
ExAC TOPMed gnomAD |
|
|
COSM4055583 COSM4055582 |
370 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754967980 | 371 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs751415271 | 372 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs2042536287 | 377 | H>Q | No | Ensembl | |
| rs369079731 | 377 | H>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373605038 | 377 | H>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1246102944 | 378 | R>Q | No | TOPMed | |
| rs575067651 | 378 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2042535499 | 380 | L>M | No |
TOPMed gnomAD |
|
| rs764834299 | 381 | A>D | No |
ExAC gnomAD |
|
| rs933799875 | 381 | A>T | No | TOPMed | |
|
rs375819705 RCV001354902 |
382 | T>A | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1355275517 | 382 | T>N | No | gnomAD | |
| rs2042534992 | 383 | A>S | No | TOPMed | |
| rs2042534856 | 383 | A>V | No |
TOPMed gnomAD |
|
| rs1595594709 | 384 | T>P | No | Ensembl | |
| rs2042534495 | 386 | T>I | No |
TOPMed gnomAD |
|
| rs1218077293 | 387 | Y>N | No | gnomAD | |
|
RCV001881682 rs2141186026 |
388 | I>V | No |
ClinVar Ensembl dbSNP |
|
| rs1368253871 | 391 | I>M | No | gnomAD | |
| rs777075734 | 391 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1276483298 | 392 | S>P | No | gnomAD | |
| rs2042533420 | 393 | K>T | No | Ensembl | |
| rs768861816 | 394 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs975730696 | 395 | Q>R | No |
TOPMed gnomAD |
|
|
COSM6142713 rs761247058 COSM6142714 |
397 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2042532500 | 398 | N>Y | No | Ensembl | |
| rs2042532215 | 399 | A>V | No | TOPMed | |
|
rs1039593484 COSM1678473 |
400 | R>C | large_intestine [Cosmic] | No |
cosmic curated TOPMed gnomAD |
|
COSM4055581 COSM4055580 rs942640629 |
400 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs942640629 | 400 | R>L | No |
TOPMed gnomAD |
|
|
COSM2217473 rs199561289 COSM4252110 |
401 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs956432408 | 404 | A>S | No |
TOPMed gnomAD |
|
| rs956432408 | 404 | A>T | No |
TOPMed gnomAD |
|
| rs1227552483 | 405 | K>R | No | TOPMed | |
| rs1475332159 | 406 | H>D | No | gnomAD | |
| TCGA novel | 406 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4867897 COSM962848 |
409 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs368263345 | 410 | K>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs749474789 | 411 | L>I | No |
ExAC gnomAD |
|
| rs1331156236 | 413 | N>S | No | TOPMed | |
| rs2042530847 | 414 | W>* | No | gnomAD | |
| rs1254911330 | 414 | W>C | No | gnomAD | |
| COSM1286460 | 416 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs986608599 | 417 | D>G | No | TOPMed | |
| rs2042530656 | 417 | D>N | No | TOPMed | |
| rs1208833861 | 419 | V>I | No | gnomAD | |
| rs1423765368 | 420 | N>I | No |
TOPMed gnomAD |
|
| rs754669251 | 421 | Q>E | No |
ExAC gnomAD |
|
| rs572788600 | 421 | Q>H | No |
1000Genomes ExAC gnomAD |
|
|
COSM4055579 COSM4055578 |
421 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779848195 | 422 | A>G | No |
ExAC gnomAD |
|
| rs1212702962 | 422 | A>P | No |
TOPMed gnomAD |
|
| rs1212702962 | 422 | A>T | No |
TOPMed gnomAD |
|
| rs779848195 | 422 | A>V | No |
ExAC gnomAD |
|
| rs757939900 | 423 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1401571603 | 424 | H>R | No | gnomAD | |
| rs2042529565 | 424 | H>Y | No | Ensembl | |
| TCGA novel | 425 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1030744947 | 426 | A>T | No |
TOPMed gnomAD |
|
| rs201349993 | 427 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs758739543 | 427 | V>L | No | Ensembl | |
| rs374075935 | 429 | Q>L | No |
ESP TOPMed gnomAD |
|
| rs1458258342 | 430 | H>P | No | gnomAD | |
| rs2042528374 | 430 | H>Q | No | gnomAD | |
| rs1458258342 | 430 | H>R | No | gnomAD | |
| rs2042528608 | 430 | H>Y | No | Ensembl | |
| TCGA novel | 432 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2042528235 | 432 | F>I | No | Ensembl | |
| rs1176295918 | 433 | I>T | No | TOPMed | |
| TCGA novel | 434 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1400931187 | 435 | V>M | No |
TOPMed gnomAD |
|
| rs1476670018 | 440 | G>R | No |
TOPMed gnomAD |
|
| rs1434897437 | 443 | T>I | No | TOPMed | |
| rs2042485004 | 444 | F>Y | No | Ensembl | |
|
COSM962846 COSM4865003 |
445 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1438476472 | 445 | E>K | No | gnomAD | |
|
COSM4783450 COSM1373513 |
449 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1467004512 | 451 | Q>P | No | gnomAD | |
| rs2042484528 | 454 | I>V | No | TOPMed | |
| rs761142691 | 457 | A>G | No |
ExAC gnomAD |
|
| rs902469654 | 457 | A>S | No |
TOPMed gnomAD |
|
| rs902469654 | 457 | A>T | No |
TOPMed gnomAD |
|
| rs2042483875 | 458 | N>I | No | gnomAD | |
| rs1239534669 | 466 | N>S | No | gnomAD | |
| rs2042260782 | 468 | H>Y | No | gnomAD | |
| rs375108002 | 472 | L>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1434730546 | 473 | E>D | No | gnomAD | |
| rs982621367 | 478 | M>L | No |
TOPMed gnomAD |
|
| rs2042259942 | 478 | M>T | No | Ensembl | |
| rs2042259797 | 481 | Q>K | No | Ensembl | |
| rs1192605843 | 485 | T>A | No | gnomAD | |
| TCGA novel | 485 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763138984 | 486 | L>I | No |
ExAC gnomAD |
|
| rs939453617 | 488 | D>G | No | TOPMed | |
| rs1735988497 | 488 | D>Y | No | Ensembl | |
| TCGA novel | 489 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745316383 | 490 | Y>C | No |
ExAC gnomAD |
|
| rs1271022249 | 494 | P>S | No | TOPMed | |
| rs761946288 | 497 | N>H | No |
ExAC gnomAD |
|
| rs1378463614 | 498 | L>F | No | TOPMed | |
| rs771911120 | 500 | E>Q | No |
ExAC gnomAD |
|
| rs1315360034 | 501 | S>L | No |
TOPMed gnomAD |
|
| rs1360785143 | 504 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs996632134 | 504 | G>V | No | TOPMed | |
| rs745659155 | 508 | L>S | No | ExAC | |
| rs1340750291 | 509 | L>M | No | gnomAD | |
| rs1340750291 | 509 | L>V | No | gnomAD | |
| TCGA novel | 518 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2042108033 | 521 | D>V | No | Ensembl | |
| rs1595577506 | 522 | T>A | No | Ensembl | |
| rs1437845969 | 522 | T>I | No | gnomAD | |
| rs1437845969 | 522 | T>N | No | gnomAD | |
| rs1238162795 | 524 | A>V | No | gnomAD | |
| rs2042107115 | 532 | L>W | No | TOPMed | |
| rs759376106 | 533 | N>D | No |
ExAC gnomAD |
|
| COSM283064 | 533 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs540811832 | 535 | C>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs540811832 | 535 | C>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1247606074 | 535 | C>W | No | gnomAD | |
| rs1284362775 | 535 | C>Y | No | gnomAD | |
| rs2042106491 | 536 | A>T | No | TOPMed | |
| rs2042106367 | 536 | A>V | No | Ensembl | |
| rs2042106251 | 537 | L>F | No | gnomAD | |
| COSM276229 | 537 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs762807088 | 539 | E>G | No |
ExAC gnomAD |
|
| rs2042106144 | 539 | E>K | No | TOPMed | |
| rs773146969 | 541 | P>L | No |
ExAC gnomAD |
|
| rs1052475148 | 542 | R>C | No | TOPMed | |
| rs769248373 | 542 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 546 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1320136208 | 546 | K>E | No |
TOPMed gnomAD |
|
| rs1391741498 | 546 | K>R | No | gnomAD | |
| rs768441698 | 547 | A>V | No |
ExAC gnomAD |
|
| rs2042105178 | 548 | F>L | No | Ensembl | |
|
COSM3502186 COSM3502185 |
551 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746616161 | 551 | Q>R | No |
ExAC TOPMed gnomAD |
|
|
COSM1562896 COSM4782641 |
554 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1179281302 | 554 | A>T | No | gnomAD | |
| rs1442120685 | 555 | D>A | No | gnomAD | |
| rs1216726125 | 560 | Q>H | No | gnomAD | |
| rs779743583 | 562 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs367622825 | 562 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs980664753 | 563 | G>A | No |
TOPMed gnomAD |
|
| rs980664753 | 563 | G>E | No |
TOPMed gnomAD |
|
| rs2042036037 | 564 | F>C | No | gnomAD | |
|
COSM962843 COSM4864087 |
565 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772384232 | 566 | E>K | No |
ExAC gnomAD |
|
| TCGA novel | 569 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2042035488 | 571 | T>A | No |
TOPMed gnomAD |
|
| rs2042035219 | 571 | T>N | No | TOPMed | |
| rs757786338 | 572 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs971624559 | 575 | E>K | No | TOPMed | |
| rs371220531 | 576 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| COSM272990 | 576 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM962842 COSM4865747 |
577 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777838101 | 577 | I>V | No |
ExAC gnomAD |
|
|
COSM962841 rs994215149 |
580 | L>I | endometrium [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| TCGA novel | 581 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1334135743 | 583 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs756131433 | 584 | K>Q | No |
ExAC gnomAD |
|
| rs2041886828 | 586 | K>N | No | Ensembl | |
| rs774811602 | 588 | L>V | No |
ExAC gnomAD |
|
| rs2041886536 | 589 | P>S | No |
TOPMed gnomAD |
|
| rs2041886536 | 589 | P>T | No |
TOPMed gnomAD |
|
| rs2141137943 | 590 | E>* | No | Ensembl | |
| rs1596383312 | 590 | E>A | No | Ensembl | |
|
rs771220127 TCGA novel |
592 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC gnomAD NCI-TCGA |
| TCGA novel | 595 | D>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2041886026 | 595 | D>E | No | TOPMed | |
| rs2041886159 | 595 | D>N | No | TOPMed | |
| rs1472832846 | 598 | A>D | No | gnomAD | |
| rs749812335 | 598 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1472832846 | 598 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2141137877 | 599 | I>L | No | Ensembl | |
| rs1323661573 | 600 | S>C | No | TOPMed | |
| rs1196638746 | 600 | S>N | No | gnomAD | |
| rs1004910937 | 605 | T>S | No |
TOPMed gnomAD |
|
| rs1320150722 | 606 | S>C | No |
TOPMed gnomAD |
|
| rs1320150722 | 606 | S>F | No |
TOPMed gnomAD |
|
| rs1206303497 | 606 | S>P | No | gnomAD | |
| TCGA novel | 607 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1385083859 | 608 | G>R | No |
TOPMed gnomAD |
|
| rs192993048 | 609 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs192993048 | 609 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs781079093 COSM962840 |
609 | R>H | endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs781079093 | 609 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs377682500 | 610 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1300268480 | 611 | P>A | No |
TOPMed gnomAD |
|
| rs1425122771 | 611 | P>L | No |
TOPMed gnomAD |
|
| rs1456657924 | 613 | T>A | No | gnomAD | |
| rs1183806482 | 614 | R>* | No | TOPMed | |
| rs751663129 | 614 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1163629002 | 615 | T>A | No | gnomAD | |
| rs765137005 | 615 | T>I | No |
ExAC gnomAD |
|
| rs1386531517 | 616 | P>L | No |
TOPMed gnomAD |
|
| rs753894018 | 617 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2041882663 | 618 | K>T | No |
TOPMed gnomAD |
|
| rs2041882582 | 621 | K>N | No | gnomAD | |
| rs2041882408 | 622 | G>D | No | gnomAD | |
| rs764090863 | 622 | G>S | No |
ExAC gnomAD |
|
| rs1487377589 | 624 | P>Q | No |
TOPMed gnomAD |
|
| rs1487377589 | 624 | P>R | No |
TOPMed gnomAD |
|
| rs556933182 | 624 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs766899142 | 625 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs759232191 | 626 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs759232191 | 626 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs546703145 | 627 | M>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
RCV001651120 RCV000242189 CA7568876 rs16964944 VAR_056180 |
627 | M>T | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1355064311 | 628 | A>S | No | gnomAD | |
| rs1355064311 | 628 | A>T | No | gnomAD | |
| rs749652957 | 630 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs770304477 | 631 | H>R | No | ExAC | |
| rs748559386 | 632 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs2041880784 | 635 | V>A | No | TOPMed | |
| rs375656230 | 641 | N>S | No |
ESP ExAC gnomAD |
|
| TCGA novel | 644 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2041830255 | 644 | H>Y | No | gnomAD | |
| rs1359781869 | 646 | L>F | No |
TOPMed gnomAD |
|
| rs761654337 | 647 | M>R | No | Ensembl | |
| TCGA novel | 648 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780989166 | 651 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs2041829452 | 652 | A>T | No |
TOPMed gnomAD |
|
| rs1194720047 | 653 | T>A | No | gnomAD | |
| rs2041829173 | 653 | T>S | No | Ensembl | |
| rs1395308887 | 654 | T>A | No |
TOPMed gnomAD |
|
|
rs754560204 COSM1216407 |
654 | T>I | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs754560204 | 654 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1488069903 | 655 | P>A | No | gnomAD | |
| rs2041828243 | 655 | P>R | No | gnomAD | |
| rs1567073251 | 656 | H>R | No | Ensembl | |
| rs369831881 | 656 | H>Y | No |
ESP TOPMed gnomAD |
|
| rs765831346 | 657 | Y>C | No |
ExAC gnomAD |
|
| TCGA novel | 657 | Y>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1393726283 | 657 | Y>H | No | gnomAD | |
|
COSM962839 rs762594836 COSM4871294 |
659 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1470856229 | 662 | K>T | No | gnomAD | |
| rs750018076 | 667 | K>T | No |
ExAC gnomAD |
|
|
rs765688942 TCGA novel |
668 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC TOPMed gnomAD NCI-TCGA |
| rs777233118 | 671 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
COSM6006131 COSM6006130 |
673 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2041641875 | 678 | V>M | No | TOPMed | |
| TCGA novel | 681 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2041641419 | 682 | R>K | No | TOPMed | |
|
COSM962838 COSM4866693 |
688 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749481784 | 690 | I>L | No |
ExAC gnomAD |
|
| rs776290728 | 690 | I>N | No |
ExAC gnomAD |
|
| rs776290728 | 690 | I>T | No |
ExAC gnomAD |
|
| rs768378080 | 691 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs1487216919 | 691 | R>Q | No | TOPMed | |
| rs1442744535 | 692 | I>T | No | gnomAD | |
| rs757941774 | 694 | A>T | No |
ExAC gnomAD |
|
| rs1366384294 | 694 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1380289478 | 696 | G>R | No |
TOPMed gnomAD |
|
| rs1380289478 | 696 | G>S | No |
TOPMed gnomAD |
|
| rs2041638432 | 698 | P>H | No | Ensembl | |
|
rs753464756 COSM4811119 COSM1301227 |
699 | S>* | Variant assessed as Somatic; HIGH impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs764380103 | 700 | R>Q | No |
ExAC gnomAD |
|
|
rs988687514 COSM962836 |
700 | R>W | endometrium [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1276349016 | 702 | T>A | No |
TOPMed gnomAD |
|
| rs2041632399 | 703 | Y>F | No | TOPMed | |
| rs1202865239 | 705 | E>D | No |
TOPMed gnomAD |
|
| rs1323992181 | 706 | F>L | No | gnomAD | |
| rs964509594 | 706 | F>V | No | Ensembl | |
| rs1596374486 | 707 | F>L | No | Ensembl | |
| rs777565635 | 707 | F>S | No |
ExAC gnomAD |
|
| TCGA novel | 707 | F>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766595663 | 709 | R>C | No | Ensembl | |
| TCGA novel | 709 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781651583 | 711 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs1432906496 | 711 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1326646328 | 712 | V>I | No | TOPMed | |
| rs1190794468 | 714 | M>I | No | gnomAD | |
| TCGA novel | 714 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201737526 | 714 | M>T | No |
TOPMed gnomAD |
|
| rs752042344 | 715 | K>N | No |
ExAC gnomAD |
|
| TCGA novel | 717 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763080608 | 718 | D>E | No |
ExAC gnomAD |
|
| rs1168921931 | 719 | V>M | No |
TOPMed gnomAD |
|
| rs200906422 | 722 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs369213886 | 722 | D>G | No | Ensembl | |
| rs1298449786 | 722 | D>Y | No | TOPMed | |
| rs538049004 | 726 | T>A | No |
1000Genomes ExAC gnomAD |
|
| rs761973889 | 727 | C>Y | No |
ExAC gnomAD |
|
| rs1244521144 | 729 | N>I | No | TOPMed | |
| rs2041628711 | 732 | E>G | No |
TOPMed gnomAD |
|
| COSM459002 | 732 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775294008 | 734 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1264801304 | 736 | L>V | No |
TOPMed gnomAD |
|
| rs1246720134 | 737 | D>G | No | gnomAD | |
| rs1362238316 | 738 | K>E | No | gnomAD | |
| rs1422369427 | 738 | K>R | No |
TOPMed gnomAD |
|
| rs1275869574 | 739 | D>G | No | gnomAD | |
| rs774197659 | 739 | D>Y | No |
ExAC gnomAD |
|
|
COSM371312 rs2041424539 |
740 | K>R | lung [Cosmic] | No |
cosmic curated TOPMed |
| rs766320263 | 742 | Q>L | No |
ExAC gnomAD |
|
|
COSM962835 COSM4864899 |
743 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1443960191 | 745 | K>E | No |
TOPMed gnomAD |
|
| rs1363253630 | 745 | K>N | No |
TOPMed gnomAD |
|
| rs762753429 | 748 | I>T | No |
ExAC gnomAD |
|
| rs769367131 | 751 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs769367131 | 751 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs2041423278 | 752 | A>D | No | Ensembl | |
| rs776415086 | 753 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs966069034 | 755 | V>A | No | Ensembl | |
|
COSM4055572 COSM4055573 |
764 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2041421516 | 767 | R>K | No | TOPMed | |
| rs200851725 | 769 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1490860698 | 769 | A>V | No |
TOPMed gnomAD |
|
| rs373353461 | 771 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs185892206 | 772 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs185892206 COSM2217454 COSM4055571 |
772 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs530642063 | 772 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs754098214 | 777 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs1343311818 | 777 | I>M | No | gnomAD | |
| rs754098214 | 777 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs764371254 | 778 | R>G | No |
1000Genomes ExAC gnomAD |
|
| COSM1258562 | 778 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754785789 | 779 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs2041419177 | 780 | W>* | No | Ensembl | |
| TCGA novel | 784 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2041418996 | 784 | K>Q | No |
TOPMed gnomAD |
|
| rs751380178 | 784 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs751380178 | 784 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs2041418666 | 785 | K>Q | No | gnomAD | |
| rs369876715 | 788 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM5743688 rs375511929 COSM5743687 |
788 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs375511929 | 788 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1200376729 | 789 | M>I | No | gnomAD | |
| rs1428943157 | 789 | M>T | No | gnomAD | |
| rs373473099 | 789 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs547967692 | 790 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM190841 COSM4055570 rs764883819 |
790 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs776486257 | 792 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs776486257 | 792 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs768370695 | 792 | A>V | No |
ExAC gnomAD |
|
| rs1208542869 | 793 | A>S | No |
TOPMed gnomAD |
|
| rs1208542869 | 793 | A>T | No |
TOPMed gnomAD |
|
| rs1323237631 | 793 | A>V | No |
TOPMed gnomAD |
|
| rs2141108051 | 796 | M>V | No | Ensembl | |
| rs775956371 | 798 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs775956371 | 798 | R>T | No |
ExAC TOPMed gnomAD |
|
| rs1292830989 | 800 | V>M | No |
TOPMed gnomAD |
|
| rs779360560 | 801 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM4055569 COSM962834 rs376727710 |
801 | R>W | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2041414365 | 803 | Y>C | No |
TOPMed gnomAD |
|
| rs1432950583 | 803 | Y>H | No |
TOPMed gnomAD |
|
| rs749354573 | 804 | Q>E | No |
ExAC gnomAD |
|
| rs1405058680 | 804 | Q>R | No | gnomAD | |
| rs1176829578 | 805 | A>D | No | gnomAD | |
| rs756361137 | 806 | R>* | No |
ExAC gnomAD |
|
| rs752818645 | 806 | R>Q | No |
ExAC gnomAD |
|
| rs758240806 | 807 | C>S | No |
ExAC gnomAD |
|
| rs561321533 | 808 | Y>C | No |
1000Genomes ExAC |
|
| rs771438988 | 810 | K>E | No |
ExAC gnomAD |
|
| rs926286487 | 810 | K>R | No |
TOPMed gnomAD |
|
|
COSM1258563 COSM4055568 |
810 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777936367 | 811 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs2041360386 | 812 | L>M | No | TOPMed | |
| rs769710517 | 813 | R>C | No |
ExAC TOPMed gnomAD |
|
|
COSM4815805 rs561485938 COSM1478191 |
813 | R>H | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs561485938 COSM4055567 COSM4055566 |
813 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs376795637 | 816 | K>E | No |
ESP ExAC gnomAD |
|
| rs2041359633 | 817 | A>V | No | TOPMed | |
| rs750260355 | 819 | T>A | No |
ExAC gnomAD |
|
| rs778785222 | 819 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1339604702 | 820 | I>T | No | gnomAD | |
| rs757292498 | 821 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs1310563687 | 821 | I>T | No | gnomAD | |
| rs757292498 | 821 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs753670387 | 822 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs753670387 | 822 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2041358426 | 823 | K>Q | No | Ensembl | |
| rs763709035 | 825 | W>R | No |
ExAC gnomAD |
|
| rs760072573 | 826 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs752461164 | 826 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM701022 COSM4863132 |
826 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767142691 | 827 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1333745220 | 828 | Y>C | No |
TOPMed gnomAD |
|
| rs1333745220 | 828 | Y>F | No |
TOPMed gnomAD |
|
| rs759128918 | 829 | V>G | No |
ExAC gnomAD |
|
| rs1198062304 | 829 | V>L | No |
TOPMed gnomAD |
|
| rs1596366411 | 830 | V>A | No | Ensembl | |
|
COSM962833 RCV000886324 rs200320492 |
831 | R>C | endometrium [Cosmic] | No |
cosmic curated ClinVar ExAC TOPMed dbSNP gnomAD |
| rs771242490 | 831 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs771242490 | 831 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs763535142 | 832 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1424084873 | 832 | R>K | No | gnomAD | |
| rs773566132 | 833 | R>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs773566132 | 833 | R>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs769986720 | 834 | Y>* | No |
ExAC gnomAD |
|
| rs1165986702 | 834 | Y>C | No |
TOPMed gnomAD |
|
|
COSM4787302 COSM1373510 |
834 | Y>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2041355519 | 835 | K>E | No | Ensembl | |
| rs1363612638 | 835 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs372246980 | 837 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1219700349 | 838 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs200977210 | 839 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs200977210 | 839 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs778874967 | 840 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs757097908 | 841 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs2414145 | 842 | I>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs753854349 | 842 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs374266181 | 843 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1337818670 | 844 | L>F | No |
TOPMed gnomAD |
|
| rs1337818670 | 844 | L>V | No |
TOPMed gnomAD |
|
| rs370606839 | 847 | Y>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1366254531 | 849 | R>* | No |
TOPMed gnomAD |
|
|
rs754654495 COSM4784082 COSM1373509 |
849 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2041353144 | 853 | A>P | No | TOPMed | |
| rs2041353144 | 853 | A>S | No | TOPMed | |
| rs766775149 | 854 | R>G | No |
ExAC gnomAD |
|
| rs1159563620 | 857 | Y>C | No |
TOPMed gnomAD |
|
| rs773832206 | 858 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs773832206 | 858 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs483352686 | 858 | R>H | No |
ExAC TOPMed gnomAD |
|
|
CA228990 rs483352686 RCV000087163 |
858 | R>L | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs773832206 | 858 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs762317547 | 859 | K>M | No |
ExAC gnomAD |
|
| rs1349507835 | 860 | I>M | No | Ensembl | |
| rs1202726331 | 860 | I>V | No | gnomAD | |
| rs751248730 | 861 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs751248730 | 861 | L>V | No |
ExAC TOPMed gnomAD |
|
|
rs1212921365 RCV001355600 |
862 | R>C | No |
ClinVar TOPMed dbSNP gnomAD |
|
|
rs202202691 RCV000995361 |
862 | R>H | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs202202691 | 862 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs765598786 | 864 | H>R | No |
ExAC gnomAD |
|
| TCGA novel | 866 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1381136865 | 867 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1352336379 | 868 | I>V | No |
TOPMed gnomAD |
|
| rs1205375695 | 869 | I>T | No | Ensembl | |
| rs762251096 | 869 | I>V | No |
ExAC gnomAD |
|
| rs998222336 | 870 | Q>H | No | TOPMed | |
| rs777257254 | 871 | K>M | No |
ExAC TOPMed gnomAD |
|
| rs764309513 | 872 | R>* | No |
ExAC gnomAD |
|
| rs764309513 | 872 | R>G | No |
ExAC gnomAD |
|
| rs183036314 | 872 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs775912427 COSM213965 |
874 | R>Q | breast [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
COSM287584 rs1418196344 |
874 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs772197482 | 875 | G>C | No |
ExAC gnomAD |
|
| rs1241138650 | 875 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2041167324 | 878 | A>G | No | TOPMed | |
|
COSM3502179 COSM3502180 |
878 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs539442211 | 879 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM3386939 rs569010771 COSM3386940 |
879 | R>H | Variant assessed as Somatic; MODERATE impact. pancreas [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1400389111 | 880 | T>I | No |
TOPMed gnomAD |
|
| rs1400389111 | 880 | T>R | No |
TOPMed gnomAD |
|
| rs201410680 | 881 | H>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs748129718 | 882 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs2041166401 | 883 | K>N | No | TOPMed | |
| rs377417175 | 883 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs2041166239 | 884 | R>T | No | gnomAD | |
| rs2041166068 | 885 | S>T | No | gnomAD | |
| rs373158952 | 886 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs754546143 | 886 | M>T | No |
ExAC gnomAD |
|
| rs779476459 | 887 | H>R | No | ExAC | |
| rs758020472 | 888 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 888 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs982945296 | 889 | I>M | No |
TOPMed gnomAD |
|
|
RCV001357906 rs750046342 |
889 | I>V | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs201829722 | 890 | I>V | No | Ensembl | |
| rs768973107 | 895 | C>F | No | gnomAD | |
| rs935879909 | 895 | C>R | No | Ensembl | |
| rs768973107 | 895 | C>S | No | gnomAD | |
| rs1353496959 | 896 | F>L | No | gnomAD | |
| rs1353496959 | 896 | F>V | No | gnomAD | |
| rs1567061934 | 897 | R>K | No | gnomAD | |
| rs764683651 | 898 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs754463537 | 898 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2041162875 | 899 | M>L | No | Ensembl | |
| rs761205312 | 900 | M>T | No |
ExAC gnomAD |
|
| rs1383347238 | 900 | M>V | No | gnomAD | |
| rs1445112599 | 901 | A>T | No | gnomAD | |
| rs369459573 | 903 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs536070522 | 903 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs774607258 | 907 | K>Q | No |
ExAC gnomAD |
|
| rs2041162017 | 907 | K>R | No |
TOPMed gnomAD |
|
| rs769513672 | 909 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs776356251 | 911 | E>K | No |
ExAC gnomAD |
|
| rs1419087237 | 912 | A>V | No |
TOPMed gnomAD |
|
| rs779455603 | 913 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs779455603 | 913 | R>G | No |
ExAC TOPMed gnomAD |
|
|
rs916981993 COSM4877151 COSM4877150 |
913 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs2041160651 | 914 | S>T | No | Ensembl | |
| rs2041160383 | 917 | R>C | No | TOPMed | |
| rs1223520467 | 917 | R>H | No |
TOPMed gnomAD |
|
| rs771591005 | 918 | Y>* | No |
ExAC gnomAD |
|
| rs1288587820 | 918 | Y>C | No |
TOPMed gnomAD |
|
| rs1372236599 | 918 | Y>H | No | gnomAD | |
| TCGA novel | 919 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs964358582 | 920 | K>R | No |
TOPMed gnomAD |
|
| rs757621774 | 923 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs757621774 | 923 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs2041158459 | 925 | M>I | No | TOPMed | |
|
COSM1517459 COSM6142716 COSM6142715 rs2041158604 |
925 | M>L | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic TOPMed |
| rs2041158316 | 927 | N>S | No | TOPMed | |
| rs368948203 | 930 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs958455380 | 931 | Q>K | No |
TOPMed gnomAD |
|
| rs772551610 | 934 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs753204590 | 934 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs767539141 | 936 | V>I | No |
ExAC gnomAD |
|
|
COSM459003 COSM4828572 |
937 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2041157089 | 938 | E>K | No | gnomAD | |
| rs1479551228 | 939 | Q>* | No | gnomAD | |
|
COSM4055565 COSM4055564 |
939 | Q>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs374079932 | 941 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs766333476 | 942 | D>E | No |
ExAC gnomAD |
|
|
RCV001763834 rs758644334 |
943 | Y>C | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs750625209 | 944 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs2041041780 | 945 | C>W | No | TOPMed | |
| rs552680211 | 945 | C>Y | No |
1000Genomes gnomAD |
|
| rs2041041473 | 947 | V>M | No | Ensembl | |
| TCGA novel | 948 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763918021 | 948 | E>V | No |
ExAC gnomAD |
|
| rs2141084985 | 949 | K>R | No | Ensembl | |
| rs370409792 | 951 | T>S | No |
ESP TOPMed gnomAD |
|
| rs367831952 | 952 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM262259 | 953 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs938071067 | 954 | E>G | No | Ensembl | |
| rs1278627477 | 954 | E>K | No | gnomAD | |
| rs765277637 | 955 | G>R | No | gnomAD | |
| rs775347983 | 956 | I>M | No |
ExAC gnomAD |
|
| rs1322320080 | 956 | I>V | No | gnomAD | |
| rs2041040432 | 958 | N>K | No | TOPMed | |
| rs1405718308 | 959 | S>C | No | gnomAD | |
| rs767505457 | 961 | T>A | No |
ExAC gnomAD |
|
|
rs1343400784 COSM4871167 COSM962832 |
965 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs199883205 | 965 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199883205 | 965 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199883205 | 965 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs773942872 | 967 | D>V | No |
ExAC gnomAD |
|
| rs1277447707 | 968 | L>V | No |
TOPMed gnomAD |
|
| rs2041039045 | 969 | E>G | No | Ensembl | |
| rs1450650257 | 969 | E>K | No | gnomAD | |
| rs369665196 | 970 | R>C | No | TOPMed | |
| rs770514590 | 970 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs748997085 | 972 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1200698212 | 974 | S>I | No | gnomAD | |
| rs1255104476 | 974 | S>R | No | gnomAD | |
| rs866012862 | 975 | E>K | No | Ensembl | |
| rs772966005 | 976 | E>K | No |
ExAC gnomAD |
|
|
COSM1323948 rs373690028 |
977 | E>K | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1005760412 | 978 | A>G | No |
TOPMed gnomAD |
|
| rs1005760412 | 978 | A>V | No |
TOPMed gnomAD |
|
| rs376989831 | 980 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2141084661 | 983 | G>E | No | Ensembl | |
| rs747429181 | 984 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs747429181 | 984 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs373825708 COSM4252079 COSM1216405 |
984 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP NCI-TCGA TOPMed gnomAD |
| rs1334514866 | 987 | S>G | No | gnomAD | |
| rs780159248 | 987 | S>R | No |
ExAC gnomAD |
|
| rs758445094 | 989 | Q>K | No |
ExAC gnomAD |
|
|
rs2041035622 COSM1373508 COSM4948834 |
990 | E>D | large_intestine Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated TOPMed gnomAD NCI-TCGA Cosmic |
| rs765434171 | 992 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs765434171 | 992 | I>V | No |
ExAC TOPMed gnomAD |
|
|
COSM2217441 COSM4055561 |
993 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200975586 | 996 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200975586 | 996 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs752540878 | 996 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs759538529 | 1000 | E>V | No |
ExAC gnomAD |
|
| TCGA novel | 1001 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2041033933 | 1002 | T>S | No | TOPMed | |
| rs774132262 | 1003 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs563030400 COSM1243418 |
1003 | R>H | oesophagus [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
RCV001772912 rs563030400 |
1003 | R>L | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs371047285 | 1004 | S>L | No |
ESP TOPMed gnomAD |
|
| rs747718802 | 1008 | C>R | No |
ExAC gnomAD |
|
| rs777079963 | 1008 | C>Y | No |
ExAC gnomAD |
|
| rs1246696457 | 1009 | I>L | No |
TOPMed gnomAD |
|
| rs2041032457 | 1009 | I>S | No |
TOPMed gnomAD |
|
| rs1246696457 | 1009 | I>V | No |
TOPMed gnomAD |
|
| rs1328870250 | 1011 | E>K | No | Ensembl | |
| rs768882499 | 1012 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1026778031 | 1014 | D>G | No | gnomAD | |
|
COSM434033 COSM4814947 |
1014 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs189981926 | 1015 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs189981926 | 1015 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs772197414 | 1016 | Y>H | No |
ExAC gnomAD |
|
| rs1235286944 | 1019 | E>Q | No | TOPMed | |
| rs2041031237 | 1020 | T>I | No | TOPMed | |
| rs1301328749 | 1021 | E>K | No | gnomAD | |
| rs770893574 | 1025 | S>* | No |
ExAC gnomAD |
|
| rs770893574 | 1025 | S>L | No |
ExAC gnomAD |
|
| rs749563645 | 1026 | N>H | No |
ExAC gnomAD |
|
| rs1261493828 | 1028 | K>T | No |
TOPMed gnomAD |
|
|
COSM3816337 COSM3816336 |
1030 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM701024 COSM4859655 |
1031 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754856360 | 1032 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs754856360 | 1032 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs751411629 | 1033 | L>S | No |
ExAC TOPMed gnomAD |
|
| rs2040850197 | 1035 | K>R | No | Ensembl | |
| rs1437716313 | 1036 | Q>K | No | gnomAD | |
| rs1390431122 | 1036 | Q>R | No | gnomAD | |
| TCGA novel | 1037 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758435490 | 1040 | A>T | No |
ExAC gnomAD |
|
| rs1413503876 | 1042 | N>Y | No | gnomAD | |
| rs372637495 | 1043 | H>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs763440706 | 1044 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs577978373 COSM701025 |
1044 | R>H | lung [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs577978373 | 1044 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs763440706 | 1044 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1225621530 | 1045 | I>F | No | TOPMed | |
| rs375922608 | 1046 | V>A | No | ESP | |
| rs1466065203 | 1047 | Q>R | No | gnomAD | |
| rs772372893 | 1048 | Q>H | No |
ExAC gnomAD |
|
| rs934723671 | 1051 | E>D | No | gnomAD | |
| rs1280858745 | 1052 | M>V | No |
TOPMed gnomAD |
|
| rs1212861361 | 1054 | E>K | No | gnomAD | |
| rs766805926 | 1056 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs763447536 | 1058 | K>R | No |
ExAC gnomAD |
|
| TCGA novel | 1058 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV002024460 rs2040724317 |
1060 | L>I | No |
ClinVar TOPMed dbSNP |
|
| rs1000123517 | 1061 | V>A | No | Ensembl | |
| rs539375837 | 1064 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1349349746 | 1065 | K>R | No | gnomAD | |
| rs748423039 | 1066 | Q>L | No |
ExAC gnomAD |
|
| rs1431582045 | 1068 | E>D | No |
TOPMed gnomAD |
|
| TCGA novel | 1068 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1386062587 | 1070 | D>N | No | TOPMed | |
| rs2040722737 | 1071 | L>F | No | gnomAD | |
| rs949784458 | 1072 | N>S | No |
TOPMed gnomAD |
|
| rs772053655 | 1074 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs2040722422 | 1074 | E>Q | No | TOPMed | |
| rs778702778 | 1075 | R>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1077 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749202486 | 1081 | L>I | No |
ExAC gnomAD |
|
| rs749202486 | 1081 | L>V | No |
ExAC gnomAD |
|
| rs752412990 | 1083 | N>Y | No |
ExAC gnomAD |
|
| rs754564461 | 1086 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs751870236 | 1086 | S>N | No |
ExAC gnomAD |
|
| rs766723050 | 1087 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs779402364 | 1087 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1226192143 | 1089 | E>A | No | gnomAD | |
| rs773740638 | 1090 | E>G | No | ExAC | |
| rs2040719342 | 1091 | R>K | No | Ensembl | |
| rs761975608 | 1092 | Y>C | No |
ExAC TOPMed gnomAD |
|
|
COSM4819484 COSM4819485 |
1093 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1411100772 | 1094 | D>Y | No | gnomAD | |
|
COSM3502176 COSM3502175 |
1095 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1351078962 | 1097 | E>Q | No |
TOPMed gnomAD |
|
| rs776798660 | 1098 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs2040718214 | 1099 | M>T | No | Ensembl | |
| rs769005480 | 1100 | T>I | No |
ExAC gnomAD |
|
| rs568705950 | 1101 | L>F | No | Ensembl | |
| rs1456937067 | 1102 | M>T | No | gnomAD | |
| rs371775585 | 1102 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2040441435 | 1104 | H>N | No | TOPMed | |
|
COSM4918734 COSM4918735 |
1104 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2040441160 | 1105 | V>M | No | Ensembl | |
| rs2040440692 | 1111 | K>N | No | Ensembl | |
| rs1254926423 | 1111 | K>R | No |
TOPMed gnomAD |
|
| TCGA novel | 1112 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754003506 | 1112 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs765857985 | 1112 | R>T | No |
ExAC gnomAD |
|
| rs1596340716 | 1113 | T>A | No | TOPMed | |
| rs963128523 | 1114 | D>V | No |
TOPMed gnomAD |
|
| rs1227465050 | 1115 | S>C | No | gnomAD | |
| rs75229735 | 1115 | S>P | No | Ensembl | |
| rs2040439877 | 1116 | T>I | No | gnomAD | |
| rs1299460030 | 1117 | H>R | No | gnomAD | |
| rs191620408 | 1120 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs372239463 | 1121 | E>D | No | ESP | |
|
rs770697352 COSM434032 COSM4814735 |
1121 | E>K | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs770697352 | 1121 | E>Q | No |
ExAC gnomAD |
|
| rs2040438788 | 1123 | E>Q | No | Ensembl | |
| rs545143802 | 1124 | Y>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs545143802 | 1124 | Y>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1489589751 | 1125 | I>T | No |
TOPMed gnomAD |
|
| rs368904213 | 1125 | I>V | No |
ESP ExAC gnomAD |
|
| rs1450569565 | 1127 | S>R | No |
TOPMed gnomAD |
|
| rs777209436 | 1128 | S>F | No |
ExAC gnomAD |
|
| rs1392453672 | 1128 | S>P | No | gnomAD | |
| rs1481416964 | 1130 | I>V | No | TOPMed | |
| rs904695954 | 1131 | A>E | No |
TOPMed gnomAD |
|
| rs780699693 | 1134 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs780699693 | 1134 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1182004906 | 1135 | D>E | No | gnomAD | |
| COSM40356 | 1137 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs950891124 | 1139 | R>G | No |
TOPMed gnomAD |
|
| rs1397513496 | 1140 | T>P | No | TOPMed | |
| rs201339297 | 1141 | E>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1200031226 | 1143 | P>A | No | TOPMed | |
| rs1238692526 | 1146 | K>R | No |
TOPMed gnomAD |
|
| rs2040080320 | 1148 | V>A | No | TOPMed | |
| rs2040080486 | 1148 | V>L | No | TOPMed | |
| rs1322129761 | 1149 | P>S | No | gnomAD | |
| rs1191703727 | 1152 | M>R | No |
TOPMed gnomAD |
|
|
COSM6077731 COSM6077732 |
1153 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2141027119 | 1155 | F>L | No | Ensembl | |
| TCGA novel | 1156 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1391908102 | 1157 | K>T | No | gnomAD | |
| TCGA novel | 1159 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757666635 | 1161 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs757666635 | 1161 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs376711165 | 1161 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1472628623 | 1165 | L>V | No | gnomAD | |
| rs778410771 | 1168 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs2141027027 | 1170 | Q>E | No | Ensembl | |
| rs1450924731 | 1171 | V>L | No | gnomAD | |
| rs2040077432 | 1172 | M>V | No | Ensembl | |
| rs1267823303 | 1173 | Q>R | No | gnomAD | |
| rs763615724 | 1174 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1328517692 | 1175 | E>A | No | TOPMed | |
| rs1328517692 | 1175 | E>G | No | TOPMed | |
|
COSM3690435 rs767451086 COSM3690436 |
1178 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
RCV000899340 rs755246020 |
1178 | R>H | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs755246020 | 1178 | R>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1179 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3816332 COSM3816333 |
1181 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1229153367 | 1181 | E>G | No | gnomAD | |
| rs2040075951 | 1183 | V>A | No | TOPMed | |
| rs751755968 | 1185 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs751755968 | 1185 | R>G | No |
ExAC gnomAD |
|
| rs765053997 | 1185 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs956627266 | 1186 | S>R | No | Ensembl | |
| rs1332131182 | 1188 | A>T | No |
TOPMed gnomAD |
|
| rs539731411 | 1191 | E>G | No | 1000Genomes | |
| rs1443058006 | 1192 | E>D | No |
TOPMed gnomAD |
|
| TCGA novel | 1192 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770710774 | 1193 | R>G | No |
ExAC gnomAD |
|
| rs762653878 | 1193 | R>K | No |
ExAC gnomAD |
|
|
COSM3816330 COSM3816331 |
1193 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2040059927 | 1196 | I>M | No |
TOPMed gnomAD |
|
|
COSM470811 COSM4857854 |
1196 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770028640 | 1197 | R>* | No |
ExAC gnomAD |
|
| rs770028640 | 1197 | R>G | No |
ExAC gnomAD |
|
| rs748726139 | 1199 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs748726139 | 1199 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs781543938 | 1200 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1379300209 | 1200 | E>G | No | TOPMed | |
|
COSM4544566 COSM4544567 |
1200 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1201 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1223074867 | 1202 | E>A | No |
TOPMed gnomAD |
|
| rs769096025 | 1203 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs747099022 | 1204 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs374830068 | 1206 | L>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs374830068 | 1206 | L>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs2040057948 | 1207 | K>R | No | gnomAD | |
| rs746171056 | 1208 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs778992454 | 1208 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs757440147 | 1209 | Q>R | No |
ExAC gnomAD |
|
| TCGA novel | 1210 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs541686219 | 1211 | L>R | No |
1000Genomes ExAC gnomAD |
|
|
COSM4815546 COSM1478190 |
1213 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2039948051 | 1213 | S>L | No |
TOPMed gnomAD |
|
| rs781090646 | 1216 | K>E | No |
ExAC gnomAD |
|
| TCGA novel | 1218 | L>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2141019831 | 1221 | E>D | No | Ensembl | |
| rs930124626 | 1224 | E>K | No |
TOPMed gnomAD |
|
| rs766026322 | 1226 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs866973592 | 1226 | R>H | No |
TOPMed gnomAD |
|
| rs1226097359 | 1228 | A>T | No | gnomAD | |
| rs2039946379 | 1229 | L>F | No | Ensembl | |
| rs757840675 | 1230 | S>N | No |
ExAC gnomAD |
|
| rs1327876183 | 1230 | S>R | No | gnomAD | |
| TCGA novel | 1231 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1422410973 | 1232 | K>I | No |
TOPMed gnomAD |
|
| rs1422410973 | 1232 | K>R | No |
TOPMed gnomAD |
|
|
COSM4055559 COSM4055560 |
1233 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs750146386 | 1233 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1365382820 | 1234 | A>G | No |
TOPMed gnomAD |
|
| rs2039945585 | 1234 | A>T | No | Ensembl | |
| rs1365382820 | 1234 | A>V | No |
TOPMed gnomAD |
|
| rs2039945011 | 1236 | E>A | No |
TOPMed gnomAD |
|
| rs2039945011 | 1236 | E>G | No |
TOPMed gnomAD |
|
| rs2141019724 | 1237 | V>M | No | Ensembl | |
| rs1596328705 | 1238 | T>P | No | Ensembl | |
| rs370437966 | 1239 | A>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs370437966 | 1239 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs2039944164 | 1239 | A>V | No | Ensembl | |
| rs1456851648 | 1240 | P>S | No |
TOPMed gnomAD |
|
| rs780999907 | 1243 | P>T | No |
TOPMed gnomAD |
|
| rs1161316783 | 1244 | A>T | No |
1000Genomes gnomAD |
|
| rs1195603232 | 1245 | Y>C | No | gnomAD | |
| rs772201102 | 1246 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs772201102 | 1246 | R>P | No |
ExAC TOPMed gnomAD |
|
|
rs2141019589 RCV001752519 |
1247 | V>A | No |
ClinVar Ensembl dbSNP |
|
| rs376330241 | 1247 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1464127742 | 1249 | M>T | No | gnomAD | |
| rs774419696 | 1249 | M>V | No |
ExAC gnomAD |
|
| rs2039942032 | 1250 | E>K | No | TOPMed | |
| rs2039941163 | 1255 | V>A | No | gnomAD | |
| rs926487258 | 1255 | V>M | No | Ensembl | |
| rs919207858 | 1257 | E>K | No |
TOPMed gnomAD |
|
| rs781177187 | 1262 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs754788532 | 1262 | R>H | No |
ExAC gnomAD |
|
| rs754788532 | 1262 | R>L | No |
ExAC gnomAD |
|
| rs1484194747 | 1263 | K>E | No |
TOPMed gnomAD |
|
| rs747008601 | 1263 | K>M | No |
ExAC gnomAD |
|
| rs1484194747 | 1263 | K>Q | No |
TOPMed gnomAD |
|
| rs780124012 | 1268 | I>T | No |
ExAC gnomAD |
|
| rs1359900869 | 1270 | R>K | No | TOPMed | |
| rs1464605020 | 1271 | S>C | No | gnomAD | |
|
COSM4786052 COSM1373506 |
1271 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1359150195 | 1272 | Q>L | No | gnomAD | |
| rs758290950 | 1273 | L>P | No |
ExAC gnomAD |
|
| rs1437049262 | 1274 | V>L | No |
TOPMed gnomAD |
|
| TCGA novel | 1275 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2039937825 | 1279 | A>D | No | gnomAD | |
| rs777833837 | 1279 | A>T | No | gnomAD | |
| rs764756469 | 1280 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs764756469 | 1280 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs2039937645 | 1280 | I>V | No | Ensembl | |
| rs756909154 | 1282 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs756909154 | 1282 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1261993244 | 1282 | P>S | No | gnomAD | |
| TCGA novel | 1282 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754427330 | 1283 | K>N | No | TOPMed | |
| rs1481997553 | 1283 | K>Q | No | gnomAD | |
| rs1236998515 | 1283 | K>R | No | Ensembl | |
| rs2039793006 | 1284 | D>N | No | TOPMed | |
| rs1461647855 | 1285 | D>G | No | gnomAD | |
| rs753550026 | 1286 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs879039494 | 1286 | K>N | No | gnomAD | |
| rs753550026 | 1286 | K>R | No |
ExAC gnomAD |
|
| rs369195695 | 1289 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs755783218 | 1289 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs2141006047 | 1293 | T>I | No | Ensembl | |
| rs2039631746 | 1293 | T>P | No | TOPMed | |
| rs1162909820 | 1294 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2039631331 | 1295 | L>V | No | TOPMed | |
| TCGA novel | 1296 | L>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2039631173 | 1297 | E>Q | No | gnomAD | |
| rs2039630835 | 1298 | D>E | No | Ensembl | |
| rs1253684582 | 1298 | D>N | No | TOPMed | |
| rs755366134 | 1299 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1362549081 | 1301 | K>R | No | gnomAD | |
| TCGA novel | 1302 | M>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763485536 | 1302 | M>V | No |
ExAC gnomAD |
|
| rs750554932 | 1304 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs983508058 | 1306 | G>D | No |
TOPMed gnomAD |
|
| rs983508058 | 1306 | G>V | No |
TOPMed gnomAD |
|
| rs1433987888 | 1307 | E>K | No | gnomAD | |
| rs2039629577 | 1308 | I>M | No | gnomAD | |
| rs369769639 | 1313 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs762167716 | 1313 | I>V | No |
ExAC gnomAD |
|
| rs1241743121 | 1314 | G>V | No | gnomAD | |
| rs950842718 | 1315 | L>V | No | Ensembl | |
| rs2039628891 | 1315 | L>W | No |
TOPMed gnomAD |
|
| rs2039628707 | 1317 | E>G | No | Ensembl | |
| rs771823878 | 1318 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs2039628294 | 1319 | N>D | No | TOPMed | |
| rs1448827892 | 1319 | N>S | No | gnomAD | |
|
CA7568242 RCV001640476 RCV000244092 rs61731219 |
1320 | R>S | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1254088119 | 1320 | R>T | No |
TOPMed gnomAD |
|
| rs1567036709 | 1325 | D>G | No | Ensembl | |
| rs764383718 | 1327 | H>R | No |
ExAC gnomAD |
|
|
COSM3816328 COSM3816329 |
1328 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1567036682 | 1331 | E>G | No | gnomAD | |
| rs1290951091 | 1332 | D>Y | No |
TOPMed gnomAD |
|
|
RCV001763295 rs2039458912 |
1334 | E>Q | No |
ClinVar Ensembl dbSNP |
|
| rs78613452 | 1338 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs78613452 | 1338 | V>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs79687584 | 1339 | Y>F | No |
ExAC gnomAD |
|
| rs773125298 | 1340 | E>D | No |
ExAC gnomAD |
|
| rs1233273553 | 1340 | E>K | No | Ensembl | |
| rs769351300 | 1342 | L>V | No | ExAC | |
| rs1270123097 | 1343 | K>Q | No | TOPMed | |
|
RCV001763833 rs747707431 |
1343 | K>R | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs775934791 | 1344 | Q>K | No | ExAC | |
| rs1329241164 | 1345 | A>V | No | gnomAD | |
| rs2039457210 | 1346 | N>D | No | TOPMed | |
| rs1411857544 | 1346 | N>K | No |
TOPMed gnomAD |
|
| rs559965743 | 1346 | N>S | No |
1000Genomes ExAC gnomAD |
|
| rs775422962 | 1347 | R>G | No | gnomAD | |
| rs547661146 | 1347 | R>K | No |
1000Genomes TOPMed gnomAD |
|
| rs1225330305 | 1347 | R>S | No |
TOPMed gnomAD |
|
| rs1368557813 | 1348 | L>F | No |
TOPMed gnomAD |
|
| rs1302354241 | 1349 | L>Q | No | gnomAD | |
| TCGA novel | 1350 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
TCGA novel rs1596313975 |
1351 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs749733587 | 1351 | S>Y | No |
ExAC gnomAD |
|
| rs1287260116 | 1353 | L>P | No | gnomAD | |
| rs1596313946 | 1355 | S>A | No | Ensembl | |
| rs2039320755 | 1357 | K>N | No | Ensembl | |
| rs1345804589 | 1357 | K>Q | No | gnomAD | |
| rs1455209908 | 1358 | R>K | No | TOPMed | |
| rs1161916927 | 1360 | H>R | No | gnomAD | |
|
COSM1301226 COSM4810619 |
1361 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2039320090 | 1362 | N>S | No | TOPMed | |
| rs2039319867 | 1363 | E>D | No | TOPMed | |
| rs753017514 | 1363 | E>K | No |
ExAC gnomAD |
|
| rs1394875492 | 1364 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs377031001 | 1365 | E>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs928150902 | 1365 | E>D | No |
TOPMed gnomAD |
|
| rs377031001 | 1365 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs377031001 | 1365 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1269013772 COSM1749214 |
1366 | A>V | urinary_tract [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1016865951 | 1367 | L>F | No |
TOPMed gnomAD |
|
| rs2039319016 | 1368 | R>C | No | Ensembl | |
|
RCV001356107 rs753717165 |
1368 | R>H | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs753717165 | 1368 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs753717165 | 1368 | R>P | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1369 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1279393077 | 1371 | I>M | No |
TOPMed gnomAD |
|
|
COSM962828 COSM4868305 |
1375 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs760117676 | 1376 | E>* | No |
ExAC gnomAD |
|
| rs1210710292 | 1376 | E>A | No | TOPMed | |
| TCGA novel | 1376 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs527716337 | 1378 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM5077247 | 1379 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1290655732 | 1380 | R>* | No | gnomAD | |
| rs1290655732 | 1380 | R>G | No | gnomAD | |
| rs759219111 | 1380 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2039316905 | 1386 | A>S | No | gnomAD | |
| rs1596313752 | 1388 | N>T | No | Ensembl | |
| rs749870708 | 1389 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1443386722 | 1390 | Q>L | No | TOPMed | |
| rs778125639 | 1392 | P>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1394 | E>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748229913 | 1396 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs1484691320 | 1396 | R>H | No | gnomAD | |
| rs781209056 | 1397 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1596313700 | 1399 | A>P | No | Ensembl | |
| TCGA novel | 1399 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751757998 | 1403 | H>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1404 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756990962 | 1407 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs756990962 | 1407 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs753611316 | 1410 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs964670972 | 1410 | N>S | No |
TOPMed gnomAD |
|
| rs964670972 | 1410 | N>T | No |
TOPMed gnomAD |
|
|
rs763946138 RCV001726955 |
1411 | E>K | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs1210719269 | 1414 | D>H | No | gnomAD | |
| rs1486205743 | 1416 | M>T | No | gnomAD | |
| rs2039126872 | 1418 | Q>R | No |
TOPMed gnomAD |
|
| rs2039126701 | 1419 | L>V | No | Ensembl | |
| rs1281120331 | 1420 | E>* | No | gnomAD | |
| TCGA novel | 1420 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1201844571 | 1423 | D>N | No |
TOPMed gnomAD |
|
| rs1337008760 | 1424 | K>E | No | gnomAD | |
| rs201575346 | 1425 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1341773336 | 1426 | V>F | No |
TOPMed gnomAD |
|
| rs1341773336 | 1426 | V>I | No |
TOPMed gnomAD |
|
| rs370782709 | 1427 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs368488059 | 1427 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1596308451 | 1430 | K>R | No | Ensembl | |
| rs2039124724 | 1432 | Q>R | No | TOPMed | |
| rs1421142198 | 1435 | V>A | No |
TOPMed gnomAD |
|
| rs375520606 | 1435 | V>I | No |
ESP TOPMed |
|
| rs1335917924 | 1438 | K>N | No | gnomAD | |
| rs2039123193 | 1438 | K>R | No | Ensembl | |
| rs1567030410 | 1440 | I>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751061571 | 1440 | I>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1440 | I>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201007049 | 1441 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs973284252 | 1442 | E>K | No |
TOPMed gnomAD |
|
| rs973284252 | 1442 | E>Q | No |
TOPMed gnomAD |
|
| TCGA novel | 1446 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1250645527 | 1447 | Q>* | No | gnomAD | |
|
COSM4928028 COSM4928027 |
1447 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1322312911 | 1448 | M>V | No | gnomAD | |
| rs895846796 | 1450 | N>K | No | TOPMed | |
| rs1274628994 | 1450 | N>S | No |
TOPMed gnomAD |
|
| rs2038838806 | 1451 | I>T | No | gnomAD | |
| rs141698310 | 1452 | S>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2038838043 | 1455 | Q>K | No | Ensembl | |
| rs761130935 | 1457 | I>M | No |
ExAC gnomAD |
|
| rs764423187 | 1457 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1020848955 | 1460 | P>T | No |
TOPMed gnomAD |
|
|
COSM3969156 COSM3969157 |
1462 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3969155 COSM3969154 |
1462 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1169465069 | 1463 | P>L | No |
TOPMed gnomAD |
|
| rs1157598301 | 1464 | V>L | No |
TOPMed gnomAD |
|
| rs2038836746 | 1465 | N>K | No | TOPMed | |
| rs550131177 | 1465 | N>S | No |
1000Genomes ExAC gnomAD |
|
| rs1381375076 | 1466 | I>V | No | gnomAD | |
| rs2038836520 | 1467 | P>A | No | Ensembl | |
| rs1321669560 | 1471 | K>R | No | Ensembl | |
| rs1441240245 | 1472 | D>E | No |
TOPMed gnomAD |
|
| rs2038836171 | 1472 | D>G | No |
TOPMed gnomAD |
|
| rs774503793 | 1474 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs2038835922 | 1474 | Q>R | No | TOPMed | |
| rs1803964710 | 1475 | G>R | No | Ensembl | |
| rs2038835557 | 1476 | M>I | No | Ensembl | |
| rs369990767 | 1478 | E>Q | No |
ESP TOPMed gnomAD |
|
| rs2038835044 | 1480 | K>E | No | Ensembl | |
| TCGA novel | 1481 | K>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM6142718 COSM6142717 |
1481 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1247903669 | 1483 | D>N | No |
TOPMed gnomAD |
|
| rs1213585712 | 1484 | E>D | No | gnomAD | |
| rs375781668 | 1488 | V>I | No |
ESP ExAC gnomAD |
|
| rs375781668 | 1488 | V>L | No |
ESP ExAC gnomAD |
|
| rs768539524 | 1489 | K>R | No |
ExAC TOPMed gnomAD |
|
|
COSM962827 COSM4864359 |
1490 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1495 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs377479179 | 1498 | R>C | No |
ESP ExAC gnomAD |
|
| rs1477212975 | 1498 | R>H | No |
TOPMed gnomAD |
|
| rs749002233 | 1500 | V>I | No |
ExAC gnomAD |
|
| rs777412090 | 1502 | V>D | No |
ExAC gnomAD |
|
| rs375254191 | 1503 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs375254191 | 1503 | N>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs781359784 | 1504 | L>M | No |
ExAC gnomAD |
|
| rs973771916 | 1505 | I>T | No |
TOPMed gnomAD |
|
| rs988976006 | 1509 | P>Q | No | Ensembl | |
| rs928841094 | 1512 | I>M | No |
TOPMed gnomAD |
|
| rs1204007548 | 1512 | I>T | No | gnomAD | |
| TCGA novel | 1517 | V>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4252047 rs373213667 COSM962826 |
1518 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs373213667 | 1518 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1518 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1355347018 | 1519 | H>R | No | gnomAD | |
|
COSM4859631 COSM701028 |
1520 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2038685422 | 1527 | Q>E | No | Ensembl | |
|
COSM4704983 COSM4704982 |
1529 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2038685208 | 1530 | R>K | No | Ensembl | |
| rs2038684812 | 1532 | L>F | No | Ensembl | |
| rs1296126185 | 1534 | T>A | No |
TOPMed gnomAD |
|
| rs1464736146 | 1534 | T>I | No | gnomAD | |
| rs183166883 | 1539 | S>G | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 1543 | V>Y | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs974195610 | 1544 | L>S | No | Ensembl | |
| rs2038683953 | 1544 | L>V | No | Ensembl | |
| rs2038427421 | 1546 | K>R | No | gnomAD | |
| rs2140936512 | 1547 | R>G | No | Ensembl | |
| rs1157793917 | 1548 | G>D | No |
TOPMed gnomAD |
|
| rs992893569 | 1549 | D>N | No |
TOPMed gnomAD |
|
| rs2038427008 | 1551 | F>V | No |
TOPMed gnomAD |
|
| rs1375544782 | 1552 | E>K | No | TOPMed | |
| rs200780801 | 1554 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1485162163 | 1558 | L>R | No | gnomAD | |
| rs1212204285 | 1558 | L>V | No | gnomAD | |
| TCGA novel | 1559 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766331108 | 1559 | S>C | No |
ExAC gnomAD |
|
| rs762825798 | 1563 | R>* | No |
ExAC gnomAD |
|
| rs772825290 | 1563 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs372394487 | 1564 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2038424715 | 1564 | F>L | No | Ensembl | |
| rs1270786712 | 1564 | F>Y | No |
TOPMed gnomAD |
|
| TCGA novel | 1565 | L>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2038424090 | 1567 | C>R | No | TOPMed | |
| rs982780154 | 1572 | S>G | No |
TOPMed gnomAD |
|
| rs761592386 | 1572 | S>N | No |
ExAC TOPMed gnomAD |
|
|
COSM4868595 COSM962825 |
1574 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1422965677 | 1576 | G>V | No | gnomAD | |
| rs760382734 | 1577 | F>L | No |
ExAC gnomAD |
|
| rs1391052390 | 1578 | M>I | No | Ensembl | |
| rs776039236 | 1578 | M>T | No |
ExAC gnomAD |
|
| COSM270715 | 1582 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772562967 | 1583 | S>Y | No |
ExAC TOPMed gnomAD |
|
|
COSM138964 rs1441544634 |
1584 | R>C | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs746447077 | 1584 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs2140930312 | 1586 | N>S | No | Ensembl | |
| rs1238419974 | 1587 | E>G | No | TOPMed | |
|
rs771406925 COSM376465 |
1592 | N>S | lung [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
COSM3377591 COSM3377590 |
1594 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs748653567 | 1596 | A>T | No | Ensembl | |
| TCGA novel | 1599 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1219218124 | 1599 | R>W | No | gnomAD | |
| rs199788926 | 1601 | V>G | No |
ExAC gnomAD |
|
| rs777777047 | 1602 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs756374551 | 1603 | S>R | No | ExAC | |
| rs779889600 | 1603 | S>R | No | ExAC | |
| rs758174824 | 1604 | D>V | No | ExAC | |
| rs1596280312 | 1607 | I>L | No | Ensembl | |
| rs1336431141 | 1608 | Q>E | No | gnomAD | |
| rs764040062 | 1608 | Q>R | No |
ExAC gnomAD |
|
|
RCV000976163 rs757170130 |
1610 | Y>F | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs775270803 | 1614 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs775270803 | 1614 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1015901825 | 1615 | R>Q | No |
TOPMed gnomAD |
|
| rs1432810247 | 1615 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1596280179 | 1616 | V>G | No | Ensembl | |
| rs1473333479 | 1616 | V>M | No | gnomAD | |
| rs767223291 | 1617 | L>S | No |
ExAC gnomAD |
|
| TCGA novel | 1618 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1596280147 | 1620 | I>T | No | Ensembl | |
| rs2038303422 | 1623 | P>L | No | Ensembl | |
| rs1456148446 | 1624 | M>T | No | gnomAD | |
| rs763433716 | 1628 | G>S | No |
ExAC gnomAD |
|
| rs2140922666 | 1629 | M>I | No | Ensembl | |
| rs2038187429 | 1629 | M>V | No | gnomAD | |
| rs2038186771 | 1631 | E>A | No | Ensembl | |
| rs2038186875 | 1631 | E>K | No | Ensembl | |
| rs2038186674 | 1632 | H>R | No | gnomAD | |
| rs1013885602 | 1634 | T>M | No |
TOPMed gnomAD |
|
| TCGA novel | 1636 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1422500964 | 1636 | Q>K | No | gnomAD | |
| rs1308807646 | 1637 | G>S | No |
TOPMed gnomAD |
|
| rs559786780 | 1638 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
TCGA novel rs2038185556 |
1640 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| TCGA novel | 1640 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4055555 COSM4055556 |
1641 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV000934043 rs200901787 |
1641 | V>L | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| TCGA novel | 1643 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768808994 | 1644 | T>R | No |
ExAC gnomAD |
|
| rs1596274669 | 1650 | T>P | No | Ensembl | |
| rs1191853928 | 1652 | S>N | No |
TOPMed gnomAD |
|
| rs1191853928 | 1652 | S>T | No |
TOPMed gnomAD |
|
| rs756082685 | 1653 | I>M | No |
ExAC gnomAD |
|
| rs752282340 | 1654 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs752282340 | 1654 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2290332 | 1655 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1361801889 | 1655 | D>G | No | gnomAD | |
| rs780727587 | 1655 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1361801889 | 1655 | D>V | No | gnomAD | |
| TCGA novel | 1655 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1455086863 | 1656 | E>D | No | gnomAD | |
| rs1596274531 | 1656 | E>V | No | Ensembl | |
| rs1357062601 | 1658 | T>I | No | gnomAD | |
| rs1174848861 | 1659 | Y>C | No | gnomAD | |
| rs962522643 | 1662 | D>H | No |
TOPMed gnomAD |
|
| rs2038182119 | 1663 | S>A | No | Ensembl | |
| rs1191446440 | 1664 | I>V | No |
TOPMed gnomAD |
|
| rs763390330 | 1666 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1406882573 | 1666 | R>W | No |
TOPMed gnomAD |
|
| rs2038181314 | 1669 | N>S | No | Ensembl | |
| rs868726502 | 1670 | S>F | No | Ensembl | |
| rs1255631085 | 1672 | H>Q | No |
TOPMed gnomAD |
|
| rs762258888 | 1672 | H>R | No |
ExAC gnomAD |
|
|
rs9282796 RCV000949585 VAR_056181 |
1673 | S>L | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs9282796 | 1673 | S>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM3420420 rs918808525 COSM3420419 |
1674 | V>D | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1256208468 | 1674 | V>I | No |
TOPMed gnomAD |
|
| rs977166419 | 1677 | Q>L | No | gnomAD | |
| rs775792965 | 1678 | H>P | No |
ExAC gnomAD |
|
| rs775792965 | 1678 | H>R | No |
ExAC gnomAD |
|
| rs2038179789 | 1678 | H>Y | No | Ensembl | |
| rs1378817610 | 1680 | M>I | No | gnomAD | |
| rs1394670963 | 1680 | M>L | No |
TOPMed gnomAD |
|
| rs1394670963 | 1680 | M>V | No |
TOPMed gnomAD |
|
| rs55901053 | 1684 | L>P | No | Ensembl | |
| rs2038179047 | 1685 | I>M | No | TOPMed | |
| rs769896407 | 1688 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs769896407 | 1688 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs143298463 | 1689 | V>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV000224692 rs143298463 CA7567943 |
1689 | V>I | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1673792949 | 1691 | Q>E | No |
TOPMed gnomAD |
|
| rs1424263949 | 1691 | Q>R | No | gnomAD | |
| rs1172360198 | 1693 | F>V | No | gnomAD | |
| rs754501060 | 1694 | Y>C | No |
ExAC gnomAD |
|
| rs780637292 | 1694 | Y>H | No |
ExAC gnomAD |
|
| rs1181388272 | 1695 | I>M | No |
TOPMed gnomAD |
|
| rs750977583 | 1696 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1446992348 | 1698 | A>T | No |
TOPMed gnomAD |
|
| rs1164841504 | 1699 | I>T | No |
TOPMed gnomAD |
|
| rs1236499563 | 1700 | T>S | No |
TOPMed gnomAD |
|
| rs1198078729 | 1704 | L>F | No | gnomAD | |
| rs758007236 | 1707 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1243205267 | 1710 | M>T | No |
TOPMed gnomAD |
|
| rs1032861712 | 1714 | S>N | No | Ensembl | |
| rs2140921848 | 1719 | I>S | No | Ensembl | |
| TCGA novel | 1720 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2038065156 | 1721 | Y>* | No | gnomAD | |
| rs757630968 | 1721 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs2140915867 | 1729 | W>* | No | Ensembl | |
| rs2038064822 | 1729 | W>G | No | TOPMed | |
| rs756527152 | 1730 | L>Q | No |
ExAC TOPMed gnomAD |
|
| rs756527152 | 1730 | L>R | No |
ExAC TOPMed gnomAD |
|
|
rs369584772 COSM4783071 COSM1373504 |
1731 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs756727033 | 1731 | R>H | No |
ExAC gnomAD |
|
| rs756727033 | 1731 | R>P | No |
ExAC gnomAD |
|
| rs2038064300 | 1732 | D>N | No | gnomAD | |
| rs1383692437 | 1733 | K>E | No | gnomAD | |
| rs774706007 | 1734 | N>H | No |
ExAC gnomAD |
|
| rs2038063885 | 1735 | L>M | No | TOPMed | |
| rs2038063672 | 1737 | N>S | No | TOPMed | |
| rs1202171950 | 1738 | S>G | No | gnomAD | |
| rs1487524515 | 1738 | S>N | No |
TOPMed gnomAD |
|
| rs1487524515 | 1738 | S>T | No |
TOPMed gnomAD |
|
| TCGA novel | 1739 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1215489861 | 1740 | A>T | No | gnomAD | |
| rs1567012887 | 1748 | I>V | No | Ensembl | |
| rs1329940411 | 1751 | A>V | No | gnomAD | |
| rs768661263 | 1754 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1268470038 | 1755 | Q>* | No | Ensembl | |
| TCGA novel | 1758 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2038061908 | 1761 | D>A | No |
TOPMed gnomAD |
|
| rs1425890819 | 1761 | D>E | No | Ensembl | |
| rs1201685288 | 1761 | D>N | No |
TOPMed gnomAD |
|
| rs1427981500 | 1762 | D>N | No |
TOPMed gnomAD |
|
| rs1169040490 | 1765 | E>D | No |
TOPMed gnomAD |
|
| rs2038061490 | 1766 | A>P | No | Ensembl | |
| rs746899428 | 1767 | I>V | No |
ExAC gnomAD |
|
| rs2038060807 | 1768 | C>S | No | gnomAD | |
|
COSM1373503 COSM4947792 |
1769 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2038060496 | 1770 | M>T | No | Ensembl | |
| rs377142501 | 1772 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1398830788 | 1773 | A>D | No | gnomAD | |
| rs1398830788 | 1773 | A>V | No | gnomAD | |
| rs2140915549 | 1774 | L>V | No | Ensembl | |
| rs190519022 | 1776 | T>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1465634909 | 1778 | Q>* | No | gnomAD | |
| rs1465634909 | 1778 | Q>K | No | gnomAD | |
| TCGA novel | 1779 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778263329 | 1779 | I>T | No |
ExAC gnomAD |
|
| rs749533763 | 1779 | I>V | No | gnomAD | |
| rs770046260 | 1780 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs770046260 | 1780 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1853329160 | 1788 | P>A | No | TOPMed | |
| TCGA novel | 1797 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV000943099 rs199675131 |
1799 | S>L | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs751712752 | 1802 | R>C | No |
ExAC gnomAD |
|
| rs1338876084 | 1802 | R>H | No | gnomAD | |
| rs201517027 | 1803 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs2037877381 | 1803 | T>I | No | TOPMed | |
| rs1244911982 | 1807 | R>C | No | gnomAD | |
| rs767265712 | 1807 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs754970527 | 1809 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs754970527 | 1809 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2140905578 | 1811 | R>G | No | Ensembl | |
| rs766285464 | 1813 | D>E | No |
ExAC gnomAD |
|
| rs1214777345 | 1813 | D>Y | No | gnomAD | |
| rs370947331 | 1816 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs376467610 | 1819 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs376467610 | 1819 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2037861165 | 1820 | D>G | No | Ensembl | |
| rs184439149 | 1821 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs761538000 | 1824 | I>V | No |
ExAC gnomAD |
|
| rs1296731918 | 1826 | P>L | No | gnomAD | |
| rs1397810527 | 1828 | T>S | No | gnomAD | |
| rs2140905455 | 1832 | N>K | No | Ensembl | |
| rs777177147 | 1833 | P>L | No |
ExAC gnomAD |
|
| rs747319127 | 1835 | S>P | No |
ExAC gnomAD |
|
| rs928993387 | 1836 | L>V | No | Ensembl | |
| rs745374294 | 1837 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs745374294 | 1837 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1480706991 | 1837 | A>V | No | gnomAD | |
|
COSM962823 COSM4867336 |
1839 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2037859015 | 1839 | E>D | No | TOPMed | |
| rs1191221756 | 1839 | E>G | No | gnomAD | |
| rs1596260147 | 1840 | T>P | No | Ensembl | |
| rs777742016 | 1841 | I>M | No | Ensembl | |
| rs370214696 | 1843 | I>M | No | ESP | |
| rs961041891 | 1846 | S>G | No |
TOPMed gnomAD |
|
| rs1016645704 | 1846 | S>T | No |
TOPMed gnomAD |
|
| rs781020390 | 1848 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs202015955 | 1854 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs202015955 | 1854 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751293557 | 1854 | R>W | No |
ExAC gnomAD |
1 associated diseases with Q9Y4I1
[MIM: 214450]: Griscelli syndrome 1 (GS1)
Rare autosomal recessive disorder that results in pigmentary dilution of the skin and hair, the presence of large clumps of pigment in hair shafts, silvery-gray hair and accumulation of melanosomes in melanocytes. GS1 patients show developmental delay, hypotonia and intellectual disability, without apparent immune abnormalities. . Note=The disease is caused by variants affecting the gene represented in this entry. Some patients who have MYO5A pathogenic variants and originally diagnosed with Griscelli syndrome 1 may rather have Elejalde syndrome. .
Without disease ID
- Rare autosomal recessive disorder that results in pigmentary dilution of the skin and hair, the presence of large clumps of pigment in hair shafts, silvery-gray hair and accumulation of melanosomes in melanocytes. GS1 patients show developmental delay, hypotonia and intellectual disability, without apparent immune abnormalities. . Note=The disease is caused by variants affecting the gene represented in this entry. Some patients who have MYO5A pathogenic variants and originally diagnosed with Griscelli syndrome 1 may rather have Elejalde syndrome. .
11 regional properties for Q9Y4I1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| binding_site | IQ motif, EF-hand binding site | 765 - 787 | IPR000048-1 |
| binding_site | IQ motif, EF-hand binding site | 788 - 810 | IPR000048-2 |
| binding_site | IQ motif, EF-hand binding site | 813 - 835 | IPR000048-3 |
| binding_site | IQ motif, EF-hand binding site | 836 - 858 | IPR000048-4 |
| binding_site | IQ motif, EF-hand binding site | 861 - 883 | IPR000048-5 |
| binding_site | IQ motif, EF-hand binding site | 884 - 906 | IPR000048-6 |
| domain | Myosin head, motor domain | 63 - 764 | IPR001609 |
| domain | Dilute domain | 1534 - 1810 | IPR002710 |
| domain | Myosin, N-terminal, SH3-like | 8 - 60 | IPR004009 |
| domain | Class V myosin, motor domain | 83 - 751 | IPR036103 |
| domain | Myosin 5a, cargo-binding domain | 1481 - 1855 | IPR037988 |
12 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| filopodium tip | The end of a filopodium distal to the body of the cell. |
| growth cone | The migrating motile tip of a growing neuron projection, where actin accumulates, and the actin cytoskeleton is the most dynamic. |
| insulin-responsive compartment | A small membrane-bounded vesicle that releases its contents by exocytosis in response to insulin stimulation; the contents are enriched in GLUT4, IRAP and VAMP2. |
| melanosome | A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
| myosin complex | A protein complex, formed of one or more myosin heavy chains plus associated light chains and other proteins, that functions as a molecular motor; uses the energy of ATP hydrolysis to move actin filaments or to move vesicles or other cargo on fixed actin filaments; has magnesium-ATPase activity and binds actin. Myosin classes are distinguished based on sequence features of the motor, or head, domain, but also have distinct tail regions that are believed to bind specific cargoes. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| microfilament motor activity | A motor activity that generates movement along a microfilament, driven by ATP hydrolysis. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| small GTPase binding | Binding to a small monomeric GTPase. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| actin filament-based movement | Movement of organelles or other particles along actin filaments, or sliding of actin filaments past each other, mediated by motor proteins. |
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| melanosome transport | The directed movement of melanosomes into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| post-Golgi vesicle-mediated transport | The directed movement of substances from the Golgi to other parts of the cell, including organelles and the plasma membrane, mediated by small transport vesicles. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| vesicle transport along actin filament | Movement of a vesicle along an actin filament, mediated by motor proteins. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
18 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q27991 | MYH10 | Myosin-10 | Bos taurus (Bovine) | SS |
| Q02440 | MYO5A | Unconventional myosin-Va | Gallus gallus (Chicken) | SS |
| Q5U651 | RASIP1 | Ras-interacting protein 1 | Homo sapiens (Human) | PR |
| P35580 | MYH10 | Myosin-10 | Homo sapiens (Human) | SS |
| B0I1T2 | MYO1G | Unconventional myosin-Ig | Homo sapiens (Human) | PR |
| Q12965 | MYO1E | Unconventional myosin-Ie | Homo sapiens (Human) | PR |
| O00160 | MYO1F | Unconventional myosin-If | Homo sapiens (Human) | PR |
| Q9UM54 | MYO6 | Unconventional myosin-VI | Homo sapiens (Human) | EV |
| Q9NQX4 | MYO5C | Unconventional myosin-Vc | Homo sapiens (Human) | SS |
| Q9ULV0 | MYO5B | Unconventional myosin-Vb | Homo sapiens (Human) | SS |
| Q3U0S6 | Rasip1 | Ras-interacting protein 1 | Mus musculus (Mouse) | PR |
| P21271 | Myo5b | Unconventional myosin-Vb | Mus musculus (Mouse) | SS |
| Q61879 | Myh10 | Myosin-10 | Mus musculus (Mouse) | SS |
| Q99104 | Myo5a | Unconventional myosin-Va | Mus musculus (Mouse) | EV |
| P70569 | Myo5b | Unconventional myosin-Vb | Rattus norvegicus (Rat) | SS |
| Q9JLT0 | Myh10 | Myosin-10 | Rattus norvegicus (Rat) | SS |
| Q9QYF3 | Myo5a | Unconventional myosin-Va | Rattus norvegicus (Rat) | SS |
| Q9M2K0 | XI-J | Myosin-16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAASELYTKF | ARVWIPDPEE | VWKSAELLKD | YKPGDKVLLL | HLEEGKDLEY | HLDPKTKELP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| HLRNPDILVG | ENDLTALSYL | HEPAVLHNLR | VRFIDSKLIY | TYCGIVLVAI | NPYEQLPIYG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EDIINAYSGQ | NMGDMDPHIF | AVAEEAYKQM | ARDERNQSII | VSGESGAGKT | VSAKYAMRYF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ATVSGSASEA | NVEEKVLASN | PIMESIGNAK | TTRNDNSSRF | GKYIEIGFDK | RYRIIGANMR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TYLLEKSRVV | FQAEEERNYH | IFYQLCASAK | LPEFKMLRLG | NADNFNYTKQ | GGSPVIEGVD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DAKEMAHTRQ | ACTLLGISES | HQMGIFRILA | GILHLGNVGF | TSRDADSCTI | PPKHEPLCIF |
| 370 | 380 | 390 | 400 | 410 | 420 |
| CELMGVDYEE | MCHWLCHRKL | ATATETYIKP | ISKLQATNAR | DALAKHIYAK | LFNWIVDNVN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| QALHSAVKQH | SFIGVLDIYG | FETFEINSFE | QFCINYANEK | LQQQFNMHVF | KLEQEEYMKE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| QIPWTLIDFY | DNQPCINLIE | SKLGILDLLD | EECKMPKGTD | DTWAQKLYNT | HLNKCALFEK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| PRLSNKAFII | QHFADKVEYQ | CEGFLEKNKD | TVFEEQIKVL | KSSKFKMLPE | LFQDDEKAIS |
| 610 | 620 | 630 | 640 | 650 | 660 |
| PTSATSSGRT | PLTRTPAKPT | KGRPGQMAKE | HKKTVGHQFR | NSLHLLMETL | NATTPHYVRC |
| 670 | 680 | 690 | 700 | 710 | 720 |
| IKPNDFKFPF | TFDEKRAVQQ | LRACGVLETI | RISAAGFPSR | WTYQEFFSRY | RVLMKQKDVL |
| 730 | 740 | 750 | 760 | 770 | 780 |
| SDRKQTCKNV | LEKLILDKDK | YQFGKTKIFF | RAGQVAYLEK | LRADKLRAAC | IRIQKTIRGW |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LLRKKYLRMR | KAAITMQRYV | RGYQARCYAK | FLRRTKAATI | IQKYWRMYVV | RRRYKIRRAA |
| 850 | 860 | 870 | 880 | 890 | 900 |
| TIVLQSYLRG | FLARNRYRKI | LREHKAVIIQ | KRVRGWLART | HYKRSMHAII | YLQCCFRRMM |
| 910 | 920 | 930 | 940 | 950 | 960 |
| AKRELKKLKI | EARSVERYKK | LHIGMENKIM | QLQRKVDEQN | KDYKCLVEKL | TNLEGIYNSE |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| TEKLRSDLER | LQLSEEEAKV | ATGRVLSLQE | EIAKLRKDLE | QTRSEKKCIE | EHADRYKQET |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| EQLVSNLKEE | NTLLKQEKEA | LNHRIVQQAK | EMTETMEKKL | VEETKQLELD | LNDERLRYQN |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| LLNEFSRLEE | RYDDLKEEMT | LMVHVPKPGH | KRTDSTHSSN | ESEYIFSSEI | AEMEDIPSRT |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| EEPSEKKVPL | DMSLFLKLQK | RVTELEQEKQ | VMQDELDRKE | EQVLRSKAKE | EERPQIRGAE |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| LEYESLKRQE | LESENKKLKN | ELNELRKALS | EKSAPEVTAP | GAPAYRVLME | QLTSVSEELD |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| VRKEEVLILR | SQLVSQKEAI | QPKDDKNTMT | DSTILLEDVQ | KMKDKGEIAQ | AYIGLKETNR |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| SSALDYHELN | EDGELWLVYE | GLKQANRLLE | SQLQSQKRSH | ENEAEALRGE | IQSLKEENNR |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| QQQLLAQNLQ | LPPEARIEAS | LQHEITRLTN | ENLDLMEQLE | KQDKTVRKLK | KQLKVFAKKI |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| GELEVGQMEN | ISPGQIIDEP | IRPVNIPRKE | KDFQGMLEYK | KEDEQKLVKN | LILELKPRGV |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| AVNLIPGLPA | YILFMCVRHA | DYLNDDQKVR | SLLTSTINSI | KKVLKKRGDD | FETVSFWLSN |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| TCRFLHCLKQ | YSGEEGFMKH | NTSRQNEHCL | TNFDLAEYRQ | VLSDLAIQIY | QQLVRVLENI |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| LQPMIVSGML | EHETIQGVSG | VKPTGLRKRT | SSIADEGTYT | LDSILRQLNS | FHSVMCQHGM |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| DPELIKQVVK | QMFYIIGAIT | LNNLLLRKDM | CSWSKGMQIR | YNVSQLEEWL | RDKNLMNSGA |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| KETLEPLIQA | AQLLQVKKKT | DDDAEAICSM | CNALTTAQIV | KVLNLYTPVN | EFEERVSVSF |
| 1810 | 1820 | 1830 | 1840 | 1850 | |
| IRTIQMRLRD | RKDSPQLLMD | AKHIFPVTFP | FNPSSLALET | IQIPASLGLG | FISRV |