Q9Y371
Gene name |
SH3GLB1 (KIAA0491, CGI-61) |
Protein name |
Endophilin-B1 |
Names |
Bax-interacting factor 1, Bif-1, SH3 domain-containing GRB2-like protein B1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:51100 |
EC number |
|
Protein Class |
|
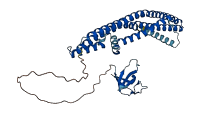
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

3 structures for Q9Y371
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6UP6 | EM | 900 A | A/B/C/D/E/F/G/H/I/J/K/L/M/N/O/P/Q/R/S/T/V/W/a/b/c/d/e/f/g/h | 1-365 | PDB |
| 6UPN | EM | 1000 A | A/B/C/D/E/F/G/H/I/J/K/L/M/N/O/P/Q/R/S/T/V/W/X/Y/a/b/c/d/e/f | 1-365 | PDB |
| AF-Q9Y371-F1 | Predicted | AlphaFoldDB |
212 variants for Q9Y371
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA341125202 rs1570397844 |
2 | N>S | No |
ClinGen Ensembl |
|
|
CA341125212 rs754775213 |
3 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs1389288016 CA341125207 |
3 | I>V | No |
ClinGen gnomAD |
|
|
CA936154 rs199927597 |
4 | M>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA936153 rs199927597 |
4 | M>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA936152 rs778761098 |
4 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs773104832 CA25925845 |
5 | D>N | No |
ClinGen Ensembl |
|
|
rs776538627 CA936155 |
7 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs1347775293 CA341125244 |
8 | V>L | No |
ClinGen gnomAD |
|
|
rs1017850515 CA25925846 |
9 | K>R | No |
ClinGen TOPMed |
|
| TCGA novel | 11 | L>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341125308 rs1176860347 |
18 | F>L | No |
ClinGen gnomAD |
|
|
rs774085197 CA25925848 |
22 | A>T | No |
ClinGen gnomAD |
|
|
CA936160 rs764542244 |
22 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs762195846 CA936162 |
23 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1307565672 CA341125373 |
26 | T>A | No |
ClinGen TOPMed |
|
|
CA936188 rs764915947 |
28 | E>Q | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 29 | K>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1467874963 CA341125418 |
32 | Q>L | No |
ClinGen gnomAD |
|
|
CA936189 rs752385047 |
33 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA341125482 rs1249889155 |
41 | H>L | No |
ClinGen gnomAD |
|
|
CA341125479 rs1558300465 |
41 | H>Y | No |
ClinGen Ensembl |
|
|
rs777505306 CA936191 |
44 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758241962 CA936190 |
44 | N>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 45 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs757330913 CA341125512 |
46 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs757330913 CA936193 |
46 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs369632434 CA936194 |
47 | S>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA341125527 rs1416623991 |
48 | K>I | No |
ClinGen TOPMed gnomAD |
|
|
CA341125530 CA341125531 rs1458106954 |
48 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
CA341125528 rs1416623991 |
48 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA341125545 rs1322953743 |
50 | E>D | No |
ClinGen gnomAD |
|
|
CA936195 rs749429791 |
51 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
CA936197 rs779312362 |
54 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA936196 rs550301160 |
54 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1434972542 CA341125572 |
55 | W>R | No |
ClinGen TOPMed |
|
|
CA936198 rs748397689 |
65 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201622706 CA936213 |
72 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA25927470 rs911801011 |
73 | A>S | No |
ClinGen TOPMed |
|
|
CA341126071 rs1208187006 |
74 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
CA341126082 rs1176678218 |
75 | I>V | No |
ClinGen TOPMed |
|
|
CA936214 rs778949424 |
78 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748392734 CA936215 |
79 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA341126154 rs1469962206 |
80 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
CA341126151 rs1230774626 |
80 | Y>D | No |
ClinGen gnomAD |
|
| TCGA novel | 84 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341126206 rs1192130798 |
87 | A>V | No |
ClinGen gnomAD |
|
|
COSM912830 CA25927471 rs778164999 |
90 | R>C | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA936217 rs778164999 |
90 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA936218 rs747420023 |
90 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs777152064 CA936221 |
91 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA936219 rs143094715 |
91 | I>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA936222 rs142378035 |
92 | N>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 93 | N>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1441282660 CA341126278 |
99 | Q>K | No |
ClinGen TOPMed gnomAD |
|
|
CA936225 rs369336536 |
99 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA25927474 rs139332321 |
100 | Y>C | No |
ClinGen ESP |
|
|
rs774149973 CA936226 |
100 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774149973 CA341126285 |
100 | Y>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1279456935 CA341126306 |
102 | I>M | No |
ClinGen gnomAD |
|
|
rs1327264404 CA341126314 |
103 | D>E | No |
ClinGen TOPMed |
|
|
CA341126307 rs1342290652 |
103 | D>N | No |
ClinGen gnomAD |
|
|
rs1196974944 CA341126325 |
105 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1196974944 CA341126326 |
105 | G>E | No |
ClinGen TOPMed gnomAD |
|
|
CA25927475 rs12035024 |
106 | T>I | No |
ClinGen TOPMed |
|
|
CA341126331 rs12035024 |
106 | T>S | No |
ClinGen TOPMed |
|
|
CA936230 rs568537654 |
111 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA341126408 rs1316076642 |
116 | N>I | No |
ClinGen gnomAD |
|
|
CA341126411 rs1352154834 |
117 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA341126458 rs766272139 |
123 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 123 | E>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs150243965 CA936250 |
124 | T>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA936254 rs757532954 |
130 | T>R | No |
ClinGen ExAC gnomAD |
|
|
CA25927798 rs990914248 |
136 | I>T | No |
ClinGen Ensembl |
|
|
CA936255 rs112687194 |
137 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA936256 rs112687194 |
137 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA936257 rs777019424 |
138 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA341126552 rs1261589668 |
138 | T>P | No |
ClinGen gnomAD |
|
|
rs748766833 CA936259 |
142 | N>H | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 143 | F>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 143 | F>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1358566715 CA341126592 |
144 | L>V | No |
ClinGen TOPMed |
|
|
rs946702002 CA25927799 |
145 | T>A | No |
ClinGen Ensembl |
|
|
CA341126603 rs1474987320 |
146 | P>A | No |
ClinGen gnomAD |
|
|
CA341126605 rs1164018187 |
146 | P>L | No |
ClinGen gnomAD |
|
| TCGA novel | 150 | F>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs778625773 CA936261 |
151 | I>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 153 | G>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1280287373 CA341126666 |
155 | Y>N | No |
ClinGen gnomAD |
|
|
CA341126675 rs1414930911 |
156 | K>E | No |
ClinGen gnomAD |
|
|
rs150004536 CA936263 |
158 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs202154654 CA936264 |
159 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs144540011 CA936265 |
159 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA936286 rs371893206 |
164 | L>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1253361628 CA341126764 |
167 | N>S | No |
ClinGen gnomAD |
|
|
CA936290 rs201518937 |
173 | D>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA341126813 rs1454513118 |
174 | A>D | No |
ClinGen TOPMed gnomAD |
|
|
CA341126815 rs1454513118 |
174 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs925229886 CA25927972 |
175 | A>T | No |
ClinGen Ensembl |
|
|
rs759734266 CA936292 |
177 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA25927973 rs1037727894 |
178 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
rs774365922 CA936295 |
180 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs761912992 CA936296 |
183 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA341126898 rs1252653068 |
187 | T>S | No |
ClinGen TOPMed |
|
|
CA936363 rs767051848 |
191 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs1570288411 CA341128005 |
192 | E>V | No |
ClinGen Ensembl |
|
|
CA341128016 rs1404546915 |
194 | E>K | No |
ClinGen Ensembl |
|
|
CA936365 rs760302020 |
197 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA936366 rs766046170 |
199 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA341128060 rs1290921745 |
200 | S>N | No |
ClinGen TOPMed |
|
|
COSM350131 rs753595065 CA936367 |
203 | D>Y | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs370892729 CA936368 |
204 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1209805059 CA341128104 |
206 | A>E | No |
ClinGen gnomAD |
|
| TCGA novel | 206 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA936369 rs778655002 |
208 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1191092462 CA341128127 |
210 | R>G | No |
ClinGen gnomAD |
|
|
CA936371 rs532928583 |
211 | L>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs753201405 CA25929003 |
214 | E>K | No |
ClinGen Ensembl |
|
|
CA936373 rs745993375 |
216 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1162603980 CA341128179 |
218 | S>N | No |
ClinGen gnomAD |
|
|
CA936375 rs375726643 |
219 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs558475875 CA25929006 |
220 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
rs749607204 CA936376 |
220 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs750491388 CA936393 |
222 | H>R | No |
ClinGen ExAC gnomAD |
|
|
TCGA novel rs1348691053 CA341128227 |
223 | H>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen gnomAD NCI-TCGA |
| TCGA novel | 224 | L>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA25929038 rs954237126 |
225 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs756276751 COSM912836 CA936394 |
225 | R>H | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1483030619 CA341128256 |
228 | N>S | No |
ClinGen gnomAD |
|
|
rs780338485 CA936395 |
230 | F>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA341128295 rs1558331225 |
233 | A>V | No |
ClinGen Ensembl |
|
|
rs1251006835 CA341128303 |
234 | Q>H | No |
ClinGen gnomAD |
|
|
rs749520521 CA936396 |
235 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA936397 rs755193646 |
238 | Y>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA341128329 rs755193646 |
238 | Y>S | No |
ClinGen ExAC gnomAD |
|
|
CA341128336 rs1454377376 |
239 | A>E | No |
ClinGen gnomAD |
|
| TCGA novel | 243 | Q>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341128375 rs1558331371 |
244 | Y>C | No |
ClinGen Ensembl |
|
|
CA25929040 rs962441730 |
245 | M>V | No |
ClinGen Ensembl |
|
|
rs1244013916 CA341128390 |
246 | L>S | No |
ClinGen TOPMed |
|
|
COSM3977942 rs965425782 CA25929041 |
249 | Q>R | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs773708467 CA936401 |
253 | G>E | No |
ClinGen ExAC gnomAD |
|
|
CA341128627 rs1443520705 |
257 | S>Y | No |
ClinGen TOPMed |
|
|
CA341128634 rs1292256271 |
258 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA936418 rs200898007 |
261 | S>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1298069442 CA341128659 |
262 | N>H | No |
ClinGen gnomAD |
|
|
rs977731111 CA25929790 |
262 | N>K | No |
ClinGen TOPMed |
|
|
rs533090731 COSM1474232 CA936419 |
264 | N>S | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
| TCGA novel | 265 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1216610042 CA341128697 |
267 | S>P | No |
ClinGen gnomAD |
|
|
rs1570303311 CA341128698 |
267 | S>Y | No |
ClinGen Ensembl |
|
|
CA341128712 rs1335181821 |
269 | T>K | No |
ClinGen gnomAD |
|
|
rs1215518202 CA341128715 |
270 | P>S | No |
ClinGen gnomAD |
|
|
rs977708561 CA25929792 |
271 | V>A | No |
ClinGen Ensembl |
|
|
CA25929793 rs924839928 |
275 | L>S | No |
ClinGen gnomAD |
|
|
CA341128743 rs1186588275 |
275 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1305391140 CA341128750 |
276 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA25929794 COSM912838 rs933506005 |
278 | A>V | Variant assessed as Somatic; impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA936424 rs769448209 |
280 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs762724218 CA936427 |
283 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs768566057 CA936428 |
283 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| TCGA novel | 285 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs553134523 CA25929795 |
285 | A>V | No |
ClinGen Ensembl |
|
|
rs1454429938 CA341128821 |
288 | S>G | No |
ClinGen gnomAD |
|
|
CA341128827 rs1290672871 |
288 | S>R | No |
ClinGen gnomAD |
|
|
rs1570303567 CA341128829 |
289 | G>S | No |
ClinGen Ensembl |
|
|
rs1338775943 CA341128835 |
290 | L>I | No |
ClinGen gnomAD |
|
|
CA341128848 rs1388018305 |
292 | I>F | No |
ClinGen TOPMed |
|
|
CA341128847 rs1388018305 |
292 | I>V | No |
ClinGen TOPMed |
|
| TCGA novel | 294 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1402694426 CA341128875 |
296 | S>F | No |
ClinGen TOPMed |
|
|
CA936429 rs774326140 |
297 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA341128880 rs774326140 |
297 | N>T | No |
ClinGen ExAC gnomAD |
|
|
rs1294089639 CA341128887 |
298 | L>R | No |
ClinGen gnomAD |
|
| TCGA novel | 301 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341128910 rs1570303658 |
301 | L>P | No |
ClinGen Ensembl |
|
|
CA936430 rs761818187 |
303 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA936431 rs767582673 |
305 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1321017409 CA341128945 |
306 | G>D | No |
ClinGen gnomAD |
|
|
CA25929797 rs924214132 |
307 | S>N | No |
ClinGen TOPMed |
|
|
rs753911559 CA341128954 |
307 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs942472279 CA25929798 |
311 | R>S | No |
ClinGen Ensembl |
|
|
CA341128987 rs1570303762 |
312 | V>G | No |
ClinGen Ensembl |
|
| TCGA novel | 313 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs138533867 CA936433 |
314 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs138533867 CA25929799 |
314 | Y>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA936434 rs562974321 |
317 | D>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs758689550 CA936436 |
320 | N>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1187286476 CA341129054 |
322 | T>I | No |
ClinGen TOPMed |
|
|
rs113079556 CA25929800 |
324 | L>S | No |
ClinGen Ensembl |
|
|
rs1420179278 CA341129092 |
328 | A>V | No |
ClinGen gnomAD |
|
|
CA936439 rs757714891 |
330 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1464793993 CA341129104 |
330 | E>G | No |
ClinGen TOPMed |
|
|
rs752866014 CA936454 |
336 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA25929875 rs1043283757 |
339 | G>E | No |
ClinGen gnomAD |
|
|
rs904322583 CA25929876 |
340 | M>I | No |
ClinGen Ensembl |
|
|
CA341129201 rs1358367325 |
343 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1358367325 CA341129203 |
343 | D>Y | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 344 | W>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341129229 rs1442596792 |
346 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
CA341129232 rs1358450575 |
347 | G>R | No |
ClinGen gnomAD |
|
|
CA341129242 rs1570305273 |
348 | E>G | No |
ClinGen Ensembl |
|
| TCGA novel | 348 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 350 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs763143617 CA936455 CA341129254 |
350 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA936456 rs764325200 |
351 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs369315549 CA936457 |
353 | K>N | No |
ClinGen ESP ExAC gnomAD |
|
|
CA341129285 rs1346443340 |
354 | G>D | No |
ClinGen gnomAD |
|
| TCGA novel | 355 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341129299 rs1570305360 |
356 | V>G | No |
ClinGen Ensembl |
|
|
CA341129334 rs1200896962 |
362 | E>K | No |
ClinGen gnomAD |
|
|
COSM1344681 rs1248562743 CA341129357 |
365 | N>S | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
No associated diseases with Q9Y371
Functions
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| autophagosome membrane | The lipid bilayer surrounding an autophagosome, a double-membrane-bounded vesicle in which endogenous cellular material is sequestered. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| mitochondrial outer membrane | The outer, i.e. cytoplasm-facing, lipid bilayer of the mitochondrial envelope. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| cadherin binding | Binding to cadherin, a type I membrane protein involved in cell adhesion. |
| identical protein binding | Binding to an identical protein or proteins. |
| lipid binding | Binding to a lipid. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
17 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| autophagic cell death | A form of programmed cell death that is accompanied by the formation of autophagosomes. Autophagic cell death is characterized by lack of chromatin condensation and massive vacuolization of the cytoplasm, with little or no uptake by phagocytic cells. |
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| cellular response to amino acid starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of amino acids. |
| cellular response to glucose starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of glucose. |
| membrane fission | A process that is carried out at the cellular level which results in the separation of a single continuous membrane into two membranes. |
| membrane organization | A process which results in the assembly, arrangement of constituent parts, or disassembly of a membrane. A membrane is a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins. |
| positive regulation of autophagosome assembly | Any process that activates or increases the frequency, rate or extent of autophagic vacuole assembly. |
| positive regulation of autophagy | Any process that activates, maintains or increases the rate of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| positive regulation of membrane tubulation | Any process that activates or increases the frequency, rate or extent of membrane tubulation. |
| positive regulation of protein targeting to mitochondrion | Any process that activates or increases the frequency, rate or extent of protein targeting to mitochondrion. |
| positive regulation of protein-containing complex assembly | Any process that activates or increases the frequency, rate or extent of protein complex assembly. |
| protein localization to vacuolar membrane | A process in which a protein is transported to, or maintained in, a location within a vacuolar membrane. |
| receptor catabolic process | The chemical reactions and pathways resulting in the breakdown of a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| regulation of cytokinesis | Any process that modulates the frequency, rate or extent of the division of the cytoplasm of a cell and its separation into two daughter cells. |
| regulation of macroautophagy | Any process that modulates the frequency, rate or extent of macroautophagy. |
| regulation of protein stability | Any process that affects the structure and integrity of a protein, altering the likelihood of its degradation or aggregation. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJA1 | SH3GL1 | Endophilin-A2 | Bos taurus (Bovine) | PR |
| Q8AXV1 | SH3GL2 | Endophilin-A1 | Gallus gallus (Chicken) | PR |
| Q5HYK7 | SH3D19 | SH3 domain-containing protein 19 | Homo sapiens (Human) | PR |
| Q99962 | SH3GL2 | Endophilin-A1 | Homo sapiens (Human) | PR |
| Q99961 | SH3GL1 | Endophilin-A2 | Homo sapiens (Human) | PR |
| Q99963 | SH3GL3 | Endophilin-A3 | Homo sapiens (Human) | PR |
| Q9Y5K6 | CD2AP | CD2-associated protein | Homo sapiens (Human) | SS |
| Q96B97 | SH3KBP1 | SH3 domain-containing kinase-binding protein 1 | Homo sapiens (Human) | EV |
| Q9JK48 | Sh3glb1 | Endophilin-B1 | Mus musculus (Mouse) | PR |
| Q62419 | Sh3gl1 | Endophilin-A2 | Mus musculus (Mouse) | PR |
| Q62420 | Sh3gl2 | Endophilin-A1 | Mus musculus (Mouse) | PR |
| Q6AYE2 | Sh3glb1 | Endophilin-B1 | Rattus norvegicus (Rat) | PR |
| O35179 | Sh3gl2 | Endophilin-A1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNIMDFNVKK | LAADAGTFLS | RAVQFTEEKL | GQAEKTELDA | HLENLLSKAE | CTKIWTEKIM |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KQTEVLLQPN | PNARIEEFVY | EKLDRKAPSR | INNPELLGQY | MIDAGTEFGP | GTAYGNALIK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| CGETQKRIGT | ADRELIQTSA | LNFLTPLRNF | IEGDYKTIAK | ERKLLQNKRL | DLDAAKTRLK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KAKAAETRNS | SEQELRITQS | EFDRQAEITR | LLLEGISSTH | AHHLRCLNDF | VEAQMTYYAQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| CYQYMLDLQK | QLGSFPSNYL | SNNNQTSVTP | VPSVLPNAIG | SSAMASTSGL | VITSPSNLSD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LKECSGSRKA | RVLYDYDAAN | STELSLLADE | VITVFSVVGM | DSDWLMGERG | NQKGKVPITY |
| LELLN |