Q9Y2H1
Gene name |
STK38L |
Protein name |
Serine/threonine-protein kinase 38-like |
Names |
NDR2 protein kinase, Nuclear Dbf2-related kinase 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:23012 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
SERINE/THREONINE-PROTEIN KINASE (PTHR24356) |
Descriptions
STK38L, called nuclear Dbf2-related kinase 2 (NDR2 protein kinase), is a subclass of the AGC family of protein kinases that are involved in the regulation of cell division and cell morphology. NDR kinase activity is regulated by phosphorylation and by the Ca2+-binding protein, S100B.
A common feature of the NDR family of kinases is an insert in the catalytic domain of 30 - 60 amino acids between subdomains VII and VIII. The insert sequence is not highly conserved but contains in all cases many positively charged amino acids. Mutations of the residues within the insert (autoinhibitory element) increase kinase activity and the binding of MOB1 or S100B to the N-terminal domain (SMA domain) of NDR induces the release of autoinhibition.
Autoinhibitory domains (AIDs)
Target domain |
90-383 (Protein kinase domain) |
Relief mechanism |
Partner binding |
Assay |
Mutagenesis experiment |
Accessory elements
230-242 (Activation loop from InterPro)
Target domain |
90-383 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
279-288 (Activation loop from InterPro)
Target domain |
90-383 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Bichsel SJ et al. (2004) "Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein", The Journal of biological chemistry, 279, 35228-35
- Stegert MR et al. (2004) "Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein", The Journal of biological chemistry, 279, 23806-12
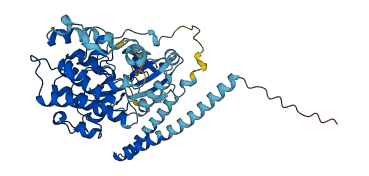
Autoinhibited structure

Activated structure

2 structures for Q9Y2H1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 5XQZ | X-ray | 210 A | C/D | 25-87 | PDB |
| AF-Q9Y2H1-F1 | Predicted | AlphaFoldDB |
207 variants for Q9Y2H1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1326997121 CA384333636 |
3 | M>V | No |
ClinGen TOPMed |
|
|
rs769405820 CA6491904 |
4 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384333655 rs1380088858 |
6 | G>R | No |
ClinGen TOPMed |
|
|
CA384333660 rs1392192254 |
6 | G>V | No |
ClinGen TOPMed |
|
|
CA384333663 rs1325173205 |
7 | T>A | No |
ClinGen gnomAD |
|
|
CA234870968 rs1000113203 |
8 | T>P | No |
ClinGen TOPMed |
|
|
rs368306630 CA6491908 |
10 | T>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1417616236 CA384333684 |
11 | F>L | No |
ClinGen TOPMed |
|
|
CA6491910 rs761216778 |
13 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA6491911 rs761216778 |
13 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA384333700 rs1349582007 |
13 | M>T | No |
ClinGen gnomAD |
|
|
rs774479388 CA6491909 |
13 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs147758452 CA6491912 |
15 | N>S | No |
ClinGen ESP ExAC gnomAD |
|
|
rs147758452 CA234870969 |
15 | N>T | No |
ClinGen ESP ExAC gnomAD |
|
|
CA6491913 rs760392409 |
17 | T>N | No |
ClinGen ExAC gnomAD |
|
|
CA384333732 rs1376483908 |
18 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
COSM693528 CA384333731 rs1213566302 |
18 | R>W | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA384333773 rs1439884455 |
24 | A>V | No |
ClinGen gnomAD |
|
|
rs766678690 CA6491914 |
25 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA234870970 rs1056825424 |
27 | T>A | No |
ClinGen gnomAD |
|
|
rs1468742613 CA384333791 |
27 | T>I | No |
ClinGen gnomAD |
|
|
rs372289940 CA6491916 |
31 | F>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA384333856 rs1461007837 |
36 | I>S | No |
ClinGen gnomAD |
|
|
CA384333860 rs1164100345 |
37 | L>* | No |
ClinGen gnomAD |
|
|
CA384333869 rs1193082021 |
38 | Q>R | No |
ClinGen TOPMed |
|
|
CA234870971 COSM938387 rs912875222 |
40 | E>G | endometrium [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA384333903 rs1411436687 |
43 | E>K | No |
ClinGen gnomAD |
|
|
rs145124460 CA234871369 |
47 | K>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA234871370 rs905792857 |
49 | L>* | No |
ClinGen Ensembl |
|
|
CA384333977 rs1175342637 |
51 | V>A | No |
ClinGen gnomAD |
|
|
rs148355950 CA6491935 |
52 | A>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs776310727 CA6491936 |
62 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1246036634 CA384334101 |
67 | R>* | No |
ClinGen TOPMed |
|
|
CA234871998 rs948046373 |
67 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs577911410 CA6491954 |
70 | H>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA234872000 rs779068249 |
71 | A>V | No |
ClinGen Ensembl |
|
|
CA6491956 rs773060776 |
72 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746501951 CA6491957 COSM430965 |
72 | R>H | breast [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA384334132 rs746501951 |
72 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384334130 rs773060776 |
72 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384334140 rs1455860370 |
73 | K>N | No |
ClinGen TOPMed |
|
|
CA234872001 rs1045151675 |
75 | T>R | No |
ClinGen TOPMed |
|
|
CA6491958 rs770482297 |
79 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA384334187 rs1215430681 |
80 | L>R | No |
ClinGen gnomAD |
|
|
rs770083865 CA6491961 |
81 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1189320275 CA384334219 |
85 | L>H | No |
ClinGen gnomAD |
|
|
rs1252214201 CA384334233 |
87 | L>F | No |
ClinGen gnomAD |
|
|
rs1591928487 CA384334237 |
88 | D>G | No |
ClinGen Ensembl |
|
|
CA6491962 rs775944298 |
88 | D>Y | No |
ClinGen ExAC |
|
|
CA384334260 rs1591928496 |
91 | E>A | No |
ClinGen Ensembl |
|
|
CA384334265 rs1591928517 |
92 | S>A | No |
ClinGen Ensembl |
|
|
CA384334289 rs1591928544 |
95 | V>G | No |
ClinGen Ensembl |
|
|
CA384334284 rs1409067668 |
95 | V>I | No |
ClinGen gnomAD |
|
|
CA6491965 rs752064286 |
97 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA6491966 rs150452352 |
98 | R>K | No |
ClinGen ESP ExAC |
|
|
rs529033839 CA6491985 |
105 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs774856321 CA6491983 |
105 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1377110922 CA384334399 |
110 | K>N | No |
ClinGen gnomAD |
|
|
rs1565551229 CA384334401 |
111 | D>H | No |
ClinGen Ensembl |
|
|
CA6491987 rs760873616 |
115 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA6491988 rs766597994 |
116 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA384334456 rs1307830246 |
119 | K>Q | No |
ClinGen gnomAD |
|
|
rs867685921 CA234872114 |
124 | S>F | No |
ClinGen Ensembl |
|
|
rs899114142 CA234872113 |
124 | S>P | No |
ClinGen Ensembl |
|
|
CA384334516 rs1272030961 |
127 | L>V | No |
ClinGen gnomAD |
|
|
rs1483641457 CA384334538 |
130 | E>* | No |
ClinGen TOPMed |
|
|
CA384334551 rs1486417528 |
131 | Q>H | No |
ClinGen gnomAD |
|
|
CA234872428 rs1024350437 |
135 | I>V | No |
ClinGen TOPMed |
|
|
rs1372876016 CA384334589 |
136 | R>* | No |
ClinGen TOPMed gnomAD |
|
|
rs757083298 CA6492014 |
136 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1041370466 CA234872429 |
140 | D>G | No |
ClinGen Ensembl |
|
|
CA384334626 rs1317351285 |
141 | I>T | No |
ClinGen TOPMed |
|
|
CA384334634 rs767429248 |
142 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384334683 rs1565553392 |
149 | W>C | No |
ClinGen Ensembl |
|
|
CA6492018 rs779445140 |
152 | K>R | No |
ClinGen ExAC |
|
|
CA384334725 rs1378160207 |
155 | Y>C | No |
ClinGen TOPMed |
|
|
CA384334729 rs1275642921 |
156 | S>G | No |
ClinGen gnomAD |
|
|
rs754620405 CA6492020 |
158 | Q>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384334748 rs754620405 |
158 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA234872431 rs896953858 |
159 | D>N | No |
ClinGen Ensembl |
|
|
CA384334762 rs113318303 |
160 | K>M | No |
ClinGen gnomAD |
|
|
CA234872432 rs113318303 |
160 | K>R | No |
ClinGen gnomAD |
|
|
rs1591936213 CA384334803 |
166 | I>N | No |
ClinGen Ensembl |
|
|
CA384334829 rs1471240770 |
169 | F>C | No |
ClinGen gnomAD |
|
|
rs1157556024 CA384334833 |
170 | L>F | No |
ClinGen TOPMed |
|
|
rs1157556024 CA384334834 |
170 | L>I | No |
ClinGen TOPMed |
|
|
rs138365234 CA234872434 |
172 | G>R | No |
ClinGen ESP |
|
|
CA6492021 rs778752373 |
172 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs1226109707 CA384334878 |
175 | M>T | No |
ClinGen gnomAD |
|
|
rs1212113825 CA384334970 |
188 | E>K | No |
ClinGen gnomAD |
|
|
rs887848651 CA234872615 |
190 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
CA234872616 rs1006329365 |
191 | Q>* | No |
ClinGen gnomAD |
|
|
CA384334991 rs1006329365 |
191 | Q>K | No |
ClinGen gnomAD |
|
|
rs142852159 CA234872617 |
191 | Q>R | No |
ClinGen ESP |
|
|
CA384335003 rs1272772856 |
192 | F>L | No |
ClinGen gnomAD |
|
|
COSM2156614 rs759763204 CA6492031 |
194 | I>V | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA6492033 rs534311579 |
199 | L>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA6492032 rs765344869 |
199 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs761591925 CA6492035 |
203 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs761591925 CA6492034 |
203 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1248786013 CA384335082 |
205 | H>Y | No |
ClinGen gnomAD |
|
|
rs1411491277 CA384335105 |
208 | G>C | No |
ClinGen gnomAD |
|
|
CA384335104 rs1411491277 |
208 | G>R | No |
ClinGen gnomAD |
|
|
rs1411491277 CA384335103 |
208 | G>S | No |
ClinGen gnomAD |
|
|
rs750304673 CA6492036 |
211 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs1034073326 CA234872618 |
212 | R>W | No |
ClinGen Ensembl |
|
|
CA6492037 rs756030339 |
218 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA6492038 rs765875075 |
220 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1282998442 CA384335209 |
223 | A>D | No |
ClinGen gnomAD |
|
|
rs1341875905 CA384335218 |
224 | K>N | No |
ClinGen gnomAD |
|
|
CA384335239 rs1297241017 |
226 | H>D | No |
ClinGen gnomAD |
|
|
CA384335238 rs1297241017 |
226 | H>N | No |
ClinGen gnomAD |
|
|
CA384335278 rs1236711543 |
231 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA384335296 rs1417705057 |
234 | L>V | No |
ClinGen TOPMed |
|
|
rs1198858335 CA384335315 |
236 | T>M | No |
ClinGen gnomAD |
|
|
rs1247126976 CA384335422 |
251 | T>I | No |
ClinGen gnomAD |
|
|
CA384335427 rs1047907482 |
252 | H>L | No |
ClinGen TOPMed |
|
|
CA234872655 rs146061018 |
252 | H>Q | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA234872654 rs1047907482 |
252 | H>R | No |
ClinGen TOPMed |
|
|
rs1267026235 CA384335433 |
253 | N>S | No |
ClinGen TOPMed |
|
|
CA6492061 rs758962045 |
254 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA6492062 rs764841037 |
256 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1565555314 CA384335448 |
256 | S>R | No |
ClinGen Ensembl |
|
|
rs752335839 CA6492063 |
257 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758144760 CA6492064 |
258 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA384335494 rs1456745082 |
260 | F>L | No |
ClinGen gnomAD |
|
|
CA6492092 rs780581159 |
262 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1224542455 CA384335587 |
273 | K>* | No |
ClinGen TOPMed |
|
|
CA384335682 rs1428780208 |
285 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1277603083 CA384335747 |
294 | V>A | No |
ClinGen TOPMed |
|
|
rs767735353 CA6492106 |
306 | W>R | No |
ClinGen ExAC gnomAD |
|
|
CA6492108 rs756523656 |
312 | I>M | No |
ClinGen ExAC gnomAD |
|
|
CA6492109 rs370057147 |
313 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1227178811 CA384335909 |
316 | M>T | No |
ClinGen gnomAD |
|
|
CA6492110 rs754414646 |
317 | L>Q | No |
ClinGen ExAC |
|
|
rs1272087052 CA384335915 |
317 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA384336058 rs1216585861 |
325 | S>F | No |
ClinGen gnomAD |
|
|
rs753220093 CA234872868 |
330 | E>G | No |
ClinGen Ensembl |
|
|
rs1478456390 CA384336121 |
332 | Y>* | No |
ClinGen gnomAD |
|
|
CA384336161 rs1235475420 |
338 | W>G | No |
ClinGen gnomAD |
|
|
rs907788113 CA234872869 |
339 | K>R | No |
ClinGen TOPMed |
|
|
CA6492135 rs780018860 COSM938391 |
343 | V>I | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed |
|
rs780018860 CA384336197 |
343 | V>L | No |
ClinGen ExAC TOPMed |
|
|
CA6492138 rs768609762 |
350 | I>T | No |
ClinGen ExAC |
|
|
CA384336243 rs1264670263 |
350 | I>V | No |
ClinGen TOPMed |
|
|
CA6492139 rs778767565 |
353 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384336273 rs1318225136 |
354 | A>V | No |
ClinGen gnomAD |
|
|
rs747683104 CA6492140 |
356 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM72837 rs1436693470 CA384336303 |
359 | L>I | ovary [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1273933914 CA384336308 |
359 | L>R | No |
ClinGen gnomAD |
|
|
CA384336304 rs1436693470 |
359 | L>V | No |
ClinGen gnomAD |
|
|
rs1416945370 CA384336336 |
362 | C>S | No |
ClinGen gnomAD |
|
|
CA6492160 rs771556609 |
363 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384336359 rs1325373369 |
365 | S>A | No |
ClinGen TOPMed |
|
|
rs1357487141 CA384336367 |
366 | E>A | No |
ClinGen gnomAD |
|
|
rs974955187 CA234873008 |
369 | I>L | No |
ClinGen TOPMed gnomAD |
|
|
rs746621114 CA6492162 |
370 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs770537407 CA6492163 |
371 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA384336423 rs1251113269 |
374 | V>A | No |
ClinGen gnomAD |
|
|
rs1482145772 CA384336429 |
375 | E>G | No |
ClinGen gnomAD |
|
|
rs373198367 CA234873009 |
379 | G>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA6492164 rs370464943 |
379 | G>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs373198367 CA6492165 |
379 | G>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA384336460 rs1472997308 |
380 | H>D | No |
ClinGen gnomAD |
|
|
rs770122828 CA384336471 |
381 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6492166 rs770122828 |
381 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763392599 CA6492168 |
383 | F>S | No |
ClinGen ExAC gnomAD |
|
|
rs763392599 CA6492169 |
383 | F>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1191103152 CA384336508 |
387 | D>N | No |
ClinGen TOPMed |
|
|
CA384336527 rs1369369010 |
389 | E>G | No |
ClinGen gnomAD |
|
|
rs148477448 CA6492170 |
391 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA384336547 rs1591947927 |
392 | R>K | No |
ClinGen Ensembl |
|
|
CA384336682 rs1235703958 |
393 | E>K | No |
ClinGen gnomAD |
|
|
CA6492191 rs773421696 |
396 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1344459477 CA384336710 |
397 | A>S | No |
ClinGen gnomAD |
|
|
rs1007662676 CA234873249 |
398 | I>V | No |
ClinGen Ensembl |
|
|
CA384336727 rs1199775474 |
400 | I>L | No |
ClinGen gnomAD |
|
|
rs760825755 CA6492192 |
400 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs1261252858 CA384336730 |
400 | I>T | No |
ClinGen gnomAD |
|
|
CA384336728 rs1199775474 |
400 | I>V | No |
ClinGen gnomAD |
|
|
CA6492193 rs765046515 |
403 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1247184826 CA384336780 |
407 | D>A | No |
ClinGen gnomAD |
|
|
rs1247184826 CA384336781 |
407 | D>G | No |
ClinGen gnomAD |
|
|
rs1306127143 CA384336800 |
410 | N>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1426322752 CA384336812 |
411 | F>L | No |
ClinGen gnomAD |
|
|
CA384336821 rs1419089559 |
413 | D>N | No |
ClinGen TOPMed |
|
|
CA6492195 rs752407589 |
416 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1477434761 CA384336853 |
417 | S>Y | No |
ClinGen gnomAD |
|
|
CA384336916 rs1289164996 |
425 | N>D | No |
ClinGen TOPMed |
|
|
rs1390210241 CA384336923 |
426 | T>A | No |
ClinGen gnomAD |
|
|
CA6492211 rs191809701 |
426 | T>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA384336947 rs1309886488 |
429 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA384336960 rs1399195893 |
431 | Y>C | No |
ClinGen TOPMed |
|
|
CA384336968 rs1338106126 |
432 | K>R | No |
ClinGen TOPMed |
|
|
CA384337036 rs1246799003 |
441 | Y>F | No |
ClinGen gnomAD |
|
|
CA6492218 rs751440994 |
445 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6492217 rs751440994 |
445 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6492222 rs779567614 |
452 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA234873252 rs958417948 |
452 | R>H | No |
ClinGen Ensembl |
|
|
CA6492223 rs753543098 |
454 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA234873253 rs956455616 |
455 | I>M | No |
ClinGen TOPMed |
|
|
rs755333316 CA6492224 |
455 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA384337140 rs1256620426 |
457 | T>I | No |
ClinGen Ensembl |
|
|
rs1165841350 CA384337142 |
458 | Y>H | No |
ClinGen gnomAD |
|
|
rs367624705 CA6492226 |
459 | M>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA6492225 rs374704707 |
459 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs772432092 CA6492227 |
462 | G>A | No |
ClinGen ExAC gnomAD |
No associated diseases with Q9Y2H1
5 regional properties for Q9Y2H1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 90 - 383 | IPR000719 |
| domain | AGC-kinase, C-terminal | 384 - 453 | IPR000961 |
| active_site | Serine/threonine-protein kinase, active site | 209 - 221 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 96 - 119 | IPR017441 |
| domain | Protein kinase, C-terminal | 402 - 443 | IPR017892 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24356 | SERINE/THREONINE-PROTEIN KINASE |
| PANTHER Subfamily | PTHR24356:SF160 | SERINE_THREONINE-PROTEIN KINASE 38-LIKE |
| PANTHER Protein Class |
non-receptor serine/threonine protein kinase
protein modifying enzyme |
|
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of cellular component organization | Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of cell structures, including the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope. |
28 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P53894 | CBK1 | Serine/threonine-protein kinase CBK1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| A2VDV2 | STK38 | Serine/threonine-protein kinase 38 | Bos taurus (Bovine) | SS |
| Q9NBK5 | trc | Serine/threonine-protein kinase tricornered | Drosophila melanogaster (Fruit fly) | SS |
| Q96LW2 | RSKR | Ribosomal protein S6 kinase-related protein | Homo sapiens (Human) | PR |
| O15530 | PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | Homo sapiens (Human) | EV |
| Q05513 | PRKCZ | Protein kinase C zeta type | Homo sapiens (Human) | SS |
| P41743 | PRKCI | Protein kinase C iota type | Homo sapiens (Human) | EV |
| Q16512 | PKN1 | Serine/threonine-protein kinase N1 | Homo sapiens (Human) | EV |
| Q6P5Z2 | PKN3 | Serine/threonine-protein kinase N3 | Homo sapiens (Human) | SS |
| Q16513 | PKN2 | Serine/threonine-protein kinase N2 | Homo sapiens (Human) | EV |
| P24723 | PRKCH | Protein kinase C eta type | Homo sapiens (Human) | SS |
| Q02156 | PRKCE | Protein kinase C epsilon type | Homo sapiens (Human) | SS |
| Q04759 | PRKCQ | Protein kinase C theta type | Homo sapiens (Human) | PR |
| Q05655 | PRKCD | Protein kinase C delta type | Homo sapiens (Human) | SS |
| P17252 | PRKCA | Protein kinase C alpha type | Homo sapiens (Human) | EV |
| P05129 | PRKCG | Protein kinase C gamma type | Homo sapiens (Human) | SS |
| P05771 | PRKCB | Protein kinase C beta type | Homo sapiens (Human) | SS |
| P31751 | AKT2 | RAC-beta serine/threonine-protein kinase | Homo sapiens (Human) | EV SS |
| P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| Q9Y243 | AKT3 | RAC-gamma serine/threonine-protein kinase | Homo sapiens (Human) | SS |
| Q96BR1 | SGK3 | Serine/threonine-protein kinase Sgk3 | Homo sapiens (Human) | SS |
| Q9HBY8 | SGK2 | Serine/threonine-protein kinase Sgk2 | Homo sapiens (Human) | SS |
| O00141 | SGK1 | Serine/threonine-protein kinase Sgk1 | Homo sapiens (Human) | PR |
| Q15208 | STK38 | Serine/threonine-protein kinase 38 | Homo sapiens (Human) | EV |
| Q6A1A2 | PDPK2P | Putative 3-phosphoinositide-dependent protein kinase 2 | Homo sapiens (Human) | PR |
| Q91VJ4 | Stk38 | Serine/threonine-protein kinase 38 | Mus musculus (Mouse) | SS |
| Q7TSE6 | Stk38l | Serine/threonine-protein kinase 38-like | Mus musculus (Mouse) | SS |
| Q2L6W9 | sax-1 | Serine/threonine-protein kinase sax-1 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAMTAGTTTT | FPMSNHTRER | VTVAKLTLEN | FYSNLILQHE | ERETRQKKLE | VAMEEEGLAD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EEKKLRRSQH | ARKETEFLRL | KRTRLGLDDF | ESLKVIGRGA | FGEVRLVQKK | DTGHIYAMKI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LRKSDMLEKE | QVAHIRAERD | ILVEADGAWV | VKMFYSFQDK | RNLYLIMEFL | PGGDMMTLLM |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KKDTLTEEET | QFYISETVLA | IDAIHQLGFI | HRDIKPDNLL | LDAKGHVKLS | DFGLCTGLKK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AHRTEFYRNL | THNPPSDFSF | QNMNSKRKAE | TWKKNRRQLA | YSTVGTPDYI | APEVFMQTGY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NKLCDWWSLG | VIMYEMLIGY | PPFCSETPQE | TYRKVMNWKE | TLVFPPEVPI | SEKAKDLILR |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FCIDSENRIG | NSGVEEIKGH | PFFEGVDWEH | IRERPAAIPI | EIKSIDDTSN | FDDFPESDIL |
| 430 | 440 | 450 | 460 | ||
| QPVPNTTEPD | YKSKDWVFLN | YTYKRFEGLT | QRGSIPTYMK | AGKL |