Q9Y243
Gene name |
AKT3 (PKBG) |
Protein name |
RAC-gamma serine/threonine-protein kinase |
Names |
Protein kinase Akt-3, Protein kinase B gamma, PKB gamma, RAC-PK-gamma, STK-2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:10000 |
EC number |
|
Protein Class |
|
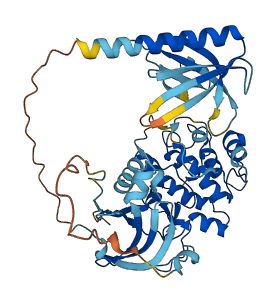
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
148-479 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
148-479 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
288-311 (Activation loop from InterPro)
Target domain |
148-405 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Huang X et al. (2003) "Crystal structure of an inactive Akt2 kinase domain", Structure (London, England : 1993), 11, 21-30
- Bichsel SJ et al. (2004) "Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein", The Journal of biological chemistry, 279, 35228-35
- Stegert MR et al. (2004) "Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein", The Journal of biological chemistry, 279, 23806-12
- Truebestein L et al. (2021) "Structure of autoinhibited Akt1 reveals mechanism of PIP(3)-mediated activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
- Lučić I et al. (2018) "Conformational sampling of membranes by Akt controls its activation and inactivation", Proceedings of the National Academy of Sciences of the United States of America, 115, E3940-E3949
- Steinberg SF (2008) "Structural basis of protein kinase C isoform function", Physiological reviews, 88, 1341-78
- Sommese RF et al. (2017) "The Role of Regulatory Domains in Maintaining Autoinhibition in the Multidomain Kinase PKCα", The Journal of biological chemistry, 292, 2873-2880
- Pears CJ et al. (1990) "Mutagenesis of the pseudosubstrate site of protein kinase C leads to activation", European journal of biochemistry, 194, 89-94
- Smith MK et al. (1990) "Specificities of autoinhibitory domain peptides for four protein kinases. Implications for intact cell studies of protein kinase function", The Journal of biological chemistry, 265, 1837-40
- Yoshinaga C et al. (1999) "Mutational analysis of the regulatory mechanism of PKN: the regulatory region of PKN contains an arachidonic acid-sensitive autoinhibitory domain", Journal of biochemistry, 126, 475-84
- Ivey RA et al. (2014) "Requirements for pseudosubstrate arginine residues during autoinhibition and phosphatidylinositol 3,4,5-(PO₄)₃-dependent activation of atypical PKC", The Journal of biological chemistry, 289, 25021-30
- Masters TA et al. (2010) "Regulation of 3-phosphoinositide-dependent protein kinase 1 activity by homodimerization in live cells", Science signaling, 3, ra78
Autoinhibited structure

Activated structure

2 structures for Q9Y243
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2X18 | X-ray | 146 A | A/B/C/D/E/F/G/H | 1-118 | PDB |
| AF-Q9Y243-F1 | Predicted | AlphaFoldDB |
277 variants for Q9Y243
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1553462330 CA345671785 RCV000623779 |
13 | Q>R | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
COSM1134860 rs397514606 VAR_065830 COSM1134859 COSM224779 CA130584 |
17 | E>K | kidney MPPH2 and melanoma; results in activation of AKT skin [Cosmic, UniProt] | Yes |
ClinGen cosmic curated UniProt Ensembl dbSNP |
|
RCV000533230 rs1553428545 CA345669292 |
75 | R>K | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001249248 rs1680073345 |
182 | E>missing | AKT3-Related Disorder [ClinVar] | Yes |
ClinVar dbSNP |
|
rs886041100 CA10602705 RCV000258932 |
183 | V>D | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000416600 VAR_069260 RCV000033036 CA130581 rs397514605 |
229 | N>S | MPPH2 Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 Megalencephaly-capillary malformation-polymicrogyria syndrome [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001364473 RCV001091228 rs1674673024 |
268 | V>A | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001289464 rs1673171921 |
321 | N>K | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1048970120 CA41022931 RCV001198092 |
414 | Y>C | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
CA345668745 RCV000557906 rs868556430 |
426 | V>L | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001172028 RCV000995482 rs1574509510 CA345668622 |
444 | I>V | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs587776935 CA130580 VAR_069261 |
465 | R>W | MPPH2; disease phenotype overlaps with megalencephaly-capillary malformation syndrome [UniProt] | Yes |
ClinGen UniProt Ensembl dbSNP |
|
rs1182345628 CA345671841 |
5 | T>A | No |
ClinGen gnomAD |
|
| rs1182345628 | 5 | T>A | No | gnomAD | |
| rs758329804 | 5 | T>I | No |
ExAC gnomAD |
|
|
rs758329804 CA1484213 |
5 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs750279631 CA1484212 |
6 | I>V | No |
ClinGen ExAC gnomAD |
|
| rs750279631 | 6 | I>V | No |
ExAC gnomAD |
|
| rs377541484 | 9 | E>A | No |
ESP ExAC gnomAD |
|
|
CA1484210 rs377541484 |
9 | E>A | No |
ClinGen ESP ExAC gnomAD |
|
| rs1553462330 | 13 | Q>R | No | Ensembl | |
| rs397514606 | 17 | E>K | No | Ensembl | |
|
CA345671412 rs1459278370 |
21 | N>Y | No |
ClinGen TOPMed |
|
| rs1459278370 | 21 | N>Y | No | TOPMed | |
| rs1199307351 | 22 | W>* | No | TOPMed | |
|
CA345671398 rs1199307351 |
22 | W>* | No |
ClinGen TOPMed |
|
| rs1261943456 | 26 | Y>F | No | gnomAD | |
|
CA345671372 rs1261943456 |
26 | Y>F | No |
ClinGen gnomAD |
|
| rs866042852 | 33 | G>V | No | Ensembl | |
|
rs866042852 CA41040445 |
33 | G>V | No |
ClinGen Ensembl |
|
|
rs1210004922 CA345671314 |
34 | S>L | No |
ClinGen gnomAD |
|
| rs1210004922 | 34 | S>L | No | gnomAD | |
| rs1346589083 | 36 | I>V | No | gnomAD | |
|
rs1346589083 CA345671304 |
36 | I>V | No |
ClinGen gnomAD |
|
|
CA345671280 rs1281393478 |
39 | K>R | No |
ClinGen gnomAD |
|
| rs1281393478 | 39 | K>R | No | gnomAD | |
|
CA345671241 rs1220113079 |
44 | D>G | No |
ClinGen gnomAD |
|
| rs1220113079 | 44 | D>G | No | gnomAD | |
| rs866698621 | 51 | L>R | No | Ensembl | |
|
rs866698621 COSM309035 CA41040442 |
51 | L>R | lung [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs1402121409 CA345671161 |
56 | V>M | No |
ClinGen gnomAD |
|
| rs1402121409 | 56 | V>M | No | gnomAD | |
| rs761062123 | 60 | Q>H | No |
ExAC gnomAD |
|
|
CA1484161 rs761062123 COSM369229 |
60 | Q>H | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
COSM1473714 CA345669353 COSM425791 rs1399248274 COSM1473715 |
66 | R>* | breast [Cosmic] | No |
ClinGen cosmic curated gnomAD |
| rs1399248274 | 66 | R>* | No | gnomAD | |
|
CA345669351 rs1328985736 |
66 | R>Q | No |
ClinGen gnomAD |
|
| rs1328985736 | 66 | R>Q | No | gnomAD | |
|
rs771617244 CA1484159 |
69 | P>S | No |
ClinGen ExAC gnomAD |
|
| rs771617244 | 69 | P>S | No |
ExAC gnomAD |
|
| rs1553428545 | 75 | R>K | No | Ensembl | |
|
rs773758145 CA1484157 |
81 | T>A | No |
ClinGen ExAC gnomAD |
|
| rs773758145 | 81 | T>A | No |
ExAC gnomAD |
|
|
CA345669238 rs1415378343 |
83 | I>V | No |
ClinGen gnomAD |
|
| rs1415378343 | 83 | I>V | No | gnomAD | |
|
CA345669211 rs1182923739 |
87 | F>L | No |
ClinGen TOPMed gnomAD |
|
| rs1182923739 | 87 | F>L | No |
TOPMed gnomAD |
|
|
rs1216797938 CA345669172 |
92 | P>Q | No |
ClinGen gnomAD |
|
| rs1216797938 | 92 | P>Q | No | gnomAD | |
|
CA1484155 rs749228367 |
94 | E>K | No |
ClinGen ExAC gnomAD |
|
| rs749228367 | 94 | E>K | No |
ExAC gnomAD |
|
| rs1024318873 | 99 | T>A | No | TOPMed | |
|
rs1024318873 CA41034427 |
99 | T>A | No |
ClinGen TOPMed |
|
| rs762440650 | 101 | A>T | No |
ExAC gnomAD |
|
|
rs762440650 CA1484138 |
101 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1300272660 CA345670756 |
105 | V>I | No |
ClinGen gnomAD |
|
| rs1300272660 | 105 | V>I | No | gnomAD | |
| rs1255579371 | 106 | A>T | No | TOPMed | |
|
CA345670750 rs1255579371 |
106 | A>T | No |
ClinGen TOPMed |
|
| rs1384888633 | 108 | R>S | No | gnomAD | |
|
rs1384888633 CA345670730 |
108 | R>S | No |
ClinGen gnomAD |
|
| rs201189866 | 111 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs201189866 CA41034426 |
111 | R>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA1484134 rs780868494 |
117 | M>I | No |
ClinGen ExAC gnomAD |
|
| rs780868494 | 117 | M>I | No |
ExAC gnomAD |
|
|
CA1484133 rs371339772 |
122 | T>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs371339772 | 122 | T>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs367585326 | 122 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
|
CA345670630 rs367585326 |
122 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs367585326 | 122 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
|
rs367585326 CA345670631 |
122 | T>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs367585326 CA1484132 |
122 | T>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs367585326 | 122 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs757469708 | 125 | I>M | No |
ExAC gnomAD |
|
|
CA1484130 rs757469708 |
125 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs200534356 CA1484131 |
125 | I>T | No |
ClinGen ExAC gnomAD |
|
| rs200534356 | 125 | I>T | No |
ExAC gnomAD |
|
|
CA41034424 RCV000493181 rs374658863 |
128 | I>M | No |
ClinGen ClinVar 1000Genomes ESP TOPMed dbSNP |
|
| rs374658863 | 128 | I>M | No |
1000Genomes ESP TOPMed |
|
| rs1487348762 | 128 | I>T | No | gnomAD | |
|
CA345670591 rs1487348762 |
128 | I>T | No |
ClinGen gnomAD |
|
| rs984642349 | 128 | I>V | No | gnomAD | |
|
CA41034425 rs984642349 |
128 | I>V | No |
ClinGen gnomAD |
|
|
CA345670585 rs1429439523 |
129 | G>A | No |
ClinGen TOPMed |
|
| rs1429439523 | 129 | G>A | No | TOPMed | |
|
rs1217463895 CA345670531 |
136 | S>F | No |
ClinGen gnomAD |
|
| rs1217463895 | 136 | S>F | No | gnomAD | |
|
rs1351256745 CA345670529 |
137 | T>A | No |
ClinGen TOPMed gnomAD |
|
| rs1351256745 | 137 | T>A | No |
TOPMed gnomAD |
|
|
CA1484128 rs202064755 |
137 | T>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs202064755 | 137 | T>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1230714185 CA345670522 |
138 | T>I | No |
ClinGen gnomAD |
|
| rs1230714185 | 138 | T>I | No | gnomAD | |
|
COSM1139119 COSM533998 rs1173790414 CA345670517 COSM1139120 |
139 | H>Y | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed |
| rs1173790414 | 139 | H>Y | No | TOPMed | |
| rs756197145 | 150 | Y>C | No |
ExAC gnomAD |
|
|
CA1484107 rs756197145 |
150 | Y>C | No |
ClinGen ExAC gnomAD |
|
| rs1426791205 | 152 | K>R | No |
TOPMed gnomAD |
|
|
CA345669881 rs1426791205 |
152 | K>R | No |
ClinGen TOPMed gnomAD |
|
| rs1085307712 | 161 | K>Q | No | Ensembl | |
|
CA345669825 RCV000490118 rs1085307712 |
161 | K>Q | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA1484104 rs755369410 |
167 | E>D | No |
ClinGen ExAC gnomAD |
|
| rs755369410 | 167 | E>D | No |
ExAC gnomAD |
|
|
CA345669765 rs1572072780 |
170 | S>C | No |
ClinGen Ensembl |
|
| rs1572072780 | 170 | S>C | No | Ensembl | |
| rs1402272180 | 171 | G>R | No | TOPMed | |
|
COSM13303 VAR_040358 rs1402272180 CA345669759 |
171 | G>R | central_nervous_system a glioblastoma multiforme sample; somatic mutation [Cosmic, UniProt] | No |
ClinGen cosmic curated UniProt TOPMed dbSNP |
| rs1472506491 | 173 | Y>C | No | gnomAD | |
|
rs1472506491 CA345669741 |
173 | Y>C | No |
ClinGen gnomAD |
|
|
rs1187260557 CA345669736 |
174 | Y>H | No |
ClinGen gnomAD |
|
| rs1187260557 | 174 | Y>H | No | gnomAD | |
| rs80155418 | 180 | K>N | No | Ensembl | |
|
CA41033285 rs80155418 |
180 | K>N | No |
ClinGen Ensembl |
|
|
rs1446319397 CA345669689 |
180 | K>R | No |
ClinGen gnomAD |
|
| rs1446319397 | 180 | K>R | No | gnomAD | |
| rs886041100 | 183 | V>D | No | Ensembl | |
| rs1243709826 | 183 | V>I | No | gnomAD | |
|
CA345669671 rs1243709826 |
183 | V>I | No |
ClinGen gnomAD |
|
| rs758066333 | 184 | I>V | No |
ExAC gnomAD |
|
|
CA1484101 rs758066333 |
184 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA345669136 rs1436246295 |
188 | D>N | No |
ClinGen gnomAD |
|
| rs1436246295 | 188 | D>N | No | gnomAD | |
| rs1343245309 | 190 | V>M | No | gnomAD | |
|
CA345669119 rs1343245309 |
190 | V>M | No |
ClinGen gnomAD |
|
| rs767433650 | 191 | A>V | No |
ExAC gnomAD |
|
|
rs767433650 CA1484073 |
191 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs887159202 CA41030717 |
194 | L>V | No |
ClinGen TOPMed gnomAD |
|
| rs887159202 | 194 | L>V | No |
TOPMed gnomAD |
|
|
rs1360057195 CA345669072 |
197 | S>N | No |
ClinGen gnomAD |
|
| rs1360057195 | 197 | S>N | No | gnomAD | |
| rs993297588 | 200 | L>F | No |
TOPMed gnomAD |
|
|
CA41030716 rs993297588 |
200 | L>F | No |
ClinGen TOPMed gnomAD |
|
| rs200428825 | 220 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
|
CA1484058 rs200428825 |
220 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1484057 rs753382585 |
221 | L>F | No |
ClinGen ExAC gnomAD |
|
| rs753382585 | 221 | L>F | No |
ExAC gnomAD |
|
| rs530590989 | 229 | N>D | No | 1000Genomes | |
|
rs530590989 CA41030554 |
229 | N>D | No |
ClinGen 1000Genomes |
|
| rs397514605 | 229 | N>S | No | Ensembl | |
| rs771134997 | 238 | S>L | No |
ExAC TOPMed gnomAD |
|
|
rs771134997 CA1484041 |
238 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs755752354 | 241 | R>L | No |
ExAC gnomAD |
|
|
rs755752354 CA1484038 |
241 | R>L | No |
ClinGen ExAC gnomAD |
|
| rs752227326 | 242 | V>L | No |
ExAC gnomAD |
|
|
rs752227326 CA1484037 |
242 | V>L | No |
ClinGen ExAC gnomAD |
|
| rs754990008 | 247 | R>C | No |
ExAC gnomAD |
|
|
CA1484035 rs754990008 |
247 | R>C | No |
ClinGen ExAC gnomAD |
|
| rs568298910 | 249 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
CA1484034 rs568298910 |
249 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1226713181 CA345671618 |
250 | F>S | No |
ClinGen gnomAD |
|
| rs1226713181 | 250 | F>S | No | gnomAD | |
| rs766197531 | 251 | Y>C | No |
ExAC TOPMed gnomAD |
|
|
CA1484033 rs766197531 |
251 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1484031 rs78293563 |
253 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| rs78293563 | 253 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs764214500 | 257 | S>C | No |
ExAC gnomAD |
|
|
rs764214500 CA1484030 |
257 | S>C | No |
ClinGen ExAC gnomAD |
|
| rs772090769 | 265 | G>R | No |
ExAC gnomAD |
|
|
CA1484027 COSM1340762 rs772090769 |
265 | G>R | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
| rs1439435818 | 270 | R>S | No | gnomAD | |
|
rs1439435818 CA345671488 |
270 | R>S | No |
ClinGen gnomAD |
|
|
CA1484003 rs752876589 |
280 | D>G | No |
ClinGen ExAC gnomAD |
|
| rs752876589 | 280 | D>G | No |
ExAC gnomAD |
|
|
rs1216018093 CA345671050 |
285 | I>M | No |
ClinGen gnomAD |
|
| rs1216018093 | 285 | I>M | No | gnomAD | |
| rs774991430 | 285 | I>V | No |
ExAC gnomAD |
|
|
rs774991430 CA1484000 |
285 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA41024856 rs778412034 |
288 | T>K | No |
ClinGen Ensembl |
|
| rs778412034 | 288 | T>K | No | Ensembl | |
| rs773474217 | 300 | A>T | No |
ExAC gnomAD |
|
|
CA1483997 COSM1501945 rs773474217 |
300 | A>T | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
| rs1168181651 | 304 | K>R | No | TOPMed | |
|
CA345670919 rs1168181651 |
304 | K>R | No |
ClinGen TOPMed |
|
|
rs1303187078 CA345670890 |
308 | G>D | No |
ClinGen gnomAD |
|
| rs1303187078 | 308 | G>D | No | gnomAD | |
| rs1064795602 | 322 | D>N | No | Ensembl | |
|
RCV000487269 rs1064795602 CA16617120 |
322 | D>N | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs1247342442 | 326 | A>T | No | TOPMed | |
|
rs1247342442 CA345670405 |
326 | A>T | No |
ClinGen TOPMed |
|
| rs773792621 | 336 | M>I | No |
ExAC gnomAD |
|
|
rs773792621 CA1483979 |
336 | M>I | No |
ClinGen ExAC gnomAD |
|
| rs763403101 | 336 | M>V | No |
ExAC gnomAD |
|
|
CA1483980 rs763403101 |
336 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA345670270 rs1019736328 |
344 | L>F | No |
ClinGen TOPMed gnomAD |
|
| rs1019736328 | 344 | L>F | No |
TOPMed gnomAD |
|
| rs1251119164 | 349 | Q>R | No | gnomAD | |
|
CA345670235 rs1251119164 |
349 | Q>R | No |
ClinGen gnomAD |
|
|
rs768388837 CA1483975 |
360 | M>T | No |
ClinGen ExAC gnomAD |
|
| rs768388837 | 360 | M>T | No |
ExAC gnomAD |
|
|
rs760280114 CA1483974 |
361 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs760280114 | 361 | E>G | No |
ExAC TOPMed gnomAD |
|
|
rs1198994311 CA345670140 |
362 | D>N | No |
ClinGen TOPMed |
|
| rs1198994311 | 362 | D>N | No | TOPMed | |
| rs1257968510 | 363 | I>T | No | TOPMed | |
|
rs1257968510 CA345670129 |
363 | I>T | No |
ClinGen TOPMed |
|
| rs771570730 | 374 | K>E | No |
ExAC gnomAD |
|
|
CA1483972 rs771570730 |
374 | K>E | No |
ClinGen ExAC gnomAD |
|
| rs1376625347 | 384 | D>G | No | TOPMed | |
|
CA345669991 rs1376625347 |
384 | D>G | No |
ClinGen TOPMed |
|
|
rs942187513 CA41022937 |
394 | D>N | No |
ClinGen Ensembl |
|
| rs942187513 | 394 | D>N | No | Ensembl | |
| rs1558604096 | 397 | K>R | No | Ensembl | |
|
rs1558604096 CA345669574 |
397 | K>R | No |
ClinGen Ensembl |
|
| rs1195814923 | 400 | M>I | No | gnomAD | |
|
rs1195814923 CA345669548 |
400 | M>I | No |
ClinGen gnomAD |
|
| rs771494754 | 401 | R>K | No |
ExAC TOPMed gnomAD |
|
|
CA1483955 rs771494754 |
401 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs910742701 | 402 | H>R | No | Ensembl | |
|
CA41022935 rs910742701 |
402 | H>R | No |
ClinGen Ensembl |
|
| rs777060434 | 402 | H>Y | No | Ensembl | |
|
CA41022936 rs777060434 |
402 | H>Y | No |
ClinGen Ensembl |
|
| rs986720230 | 409 | N>K | No | gnomAD | |
| rs986720230 | 409 | N>K | No | gnomAD | |
|
CA345669486 rs986720230 |
409 | N>K | No |
ClinGen gnomAD |
|
|
CA41022933 rs986720230 |
409 | N>K | No |
ClinGen gnomAD |
|
|
rs113943363 CA41022934 |
409 | N>S | No |
ClinGen Ensembl |
|
| rs113943363 | 409 | N>S | No | Ensembl | |
| rs769299773 | 413 | V>I | No | TOPMed | |
|
rs769299773 CA41022932 |
413 | V>I | No |
ClinGen TOPMed |
|
| rs1048970120 | 414 | Y>C | No |
TOPMed gnomAD |
|
|
rs1344843803 CA345668793 |
418 | L>P | No |
ClinGen TOPMed |
|
| rs1344843803 | 418 | L>P | No | TOPMed | |
|
rs762883936 CA1483932 |
420 | P>S | No |
ClinGen ExAC gnomAD |
|
| rs762883936 | 420 | P>S | No |
ExAC gnomAD |
|
| rs769538692 | 421 | P>A | No |
ExAC gnomAD |
|
|
CA1483930 rs769538692 |
421 | P>A | No |
ClinGen ExAC gnomAD |
|
| rs1290444154 | 424 | P>L | No | TOPMed | |
|
rs1290444154 CA345668755 |
424 | P>L | No |
ClinGen TOPMed |
|
| rs868556430 | 426 | V>I | No | Ensembl | |
|
rs868556430 CA41019244 |
426 | V>I | No |
ClinGen Ensembl |
|
| rs868556430 | 426 | V>L | No | Ensembl | |
| rs775611627 | 427 | T>I | No |
ExAC gnomAD |
|
|
rs775611627 CA1483928 |
427 | T>I | No |
ClinGen ExAC gnomAD |
|
| rs746036442 | 430 | T>I | No |
ExAC gnomAD |
|
|
rs746036442 CA1483926 |
430 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs888799070 CA41019243 |
439 | F>L | No |
ClinGen Ensembl |
|
| rs888799070 | 439 | F>L | No | Ensembl | |
| rs964744343 | 440 | T>I | No | TOPMed | |
|
rs964744343 CA41019242 |
440 | T>I | No |
ClinGen TOPMed |
|
|
CA345668638 rs1325571748 |
441 | A>V | No |
ClinGen TOPMed |
|
| rs1325571748 | 441 | A>V | No | TOPMed | |
| rs1574509510 | 444 | I>V | No | Ensembl | |
|
CA1483924 rs757338728 |
446 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs757338728 | 446 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs368205016 | 452 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
|
CA1483909 rs368205016 |
452 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1442713931 CA345668569 |
452 | Y>H | No |
ClinGen gnomAD |
|
| rs1442713931 | 452 | Y>H | No | gnomAD | |
|
rs375329841 CA1483908 |
453 | D>G | No |
ClinGen ESP ExAC gnomAD |
|
| rs375329841 | 453 | D>G | No |
ESP ExAC gnomAD |
|
|
rs1270894056 CA345668516 |
457 | M>I | No |
ClinGen gnomAD |
|
| rs1270894056 | 457 | M>I | No | gnomAD | |
|
rs1216439857 CA345668500 |
459 | C>F | No |
ClinGen gnomAD |
|
| rs1216439857 | 459 | C>F | No | gnomAD | |
| rs1197744936 | 460 | M>T | No | TOPMed | |
|
rs1197744936 CA345668495 |
460 | M>T | No |
ClinGen TOPMed |
|
| rs774657057 | 460 | M>V | No |
ExAC gnomAD |
|
|
rs774657057 CA1483906 |
460 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs771193056 CA1483905 |
461 | D>E | No |
ClinGen ExAC gnomAD |
|
| rs771193056 | 461 | D>E | No |
ExAC gnomAD |
|
|
CA1483904 rs749347952 |
462 | N>S | No |
ClinGen ExAC gnomAD |
|
| rs749347952 | 462 | N>S | No |
ExAC gnomAD |
|
| rs587776935 | 465 | R>W | No | Ensembl | |
|
rs1287560468 CA345668377 |
477 | G>R | No |
ClinGen TOPMed |
|
| rs1287560468 | 477 | G>R | No | TOPMed | |
| rs751275167 | 478 | R>Q | No |
ExAC gnomAD |
|
|
CA1483898 rs751275167 |
478 | R>Q | No |
ClinGen ExAC gnomAD |
No associated diseases with Q9Y243
7 regional properties for Q9Y243
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 501 - 786 | IPR000719 |
| domain | S-locus glycoprotein domain | 206 - 316 | IPR000858 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 502 - 770 | IPR001245 |
| domain | Bulb-type lectin domain | 24 - 174 | IPR001480 |
| domain | PAN/Apple domain | 337 - 423 | IPR003609 |
| active_site | Serine/threonine-protein kinase, active site | 622 - 634 | IPR008271 |
| domain | S-locus receptor kinase, C-terminal | 773 - 814 | IPR021820 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| brain morphogenesis | The process in which the anatomical structures of the brain are generated and organized. The brain is one of the two components of the central nervous system and is the center of thought and emotion. It is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| homeostasis of number of cells within a tissue | Any biological process involved in the maintenance of the steady-state number of cells within a population of cells in a tissue. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| mitochondrial genome maintenance | The maintenance of the structure and integrity of the mitochondrial genome; includes replication and segregation of the mitochondrial chromosome. |
| negative regulation of cellular senescence | Any process that stops, prevents or reduces the frequency, rate or extent of cellular senescence. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of angiogenesis | Any process that activates or increases angiogenesis. |
| positive regulation of artery morphogenesis | Any process that activates or increases the frequency, rate or extent of artery morphogenesis. |
| positive regulation of blood vessel endothelial cell migration | Any process that activates or increases the frequency, rate or extent of the migration of the endothelial cells of blood vessels. |
| positive regulation of cell migration involved in sprouting angiogenesis | Any process that increases the frequency, rate or extent of cell migration involved in sprouting angiogenesis. Cell migration involved in sprouting angiogenesis is the orderly movement of endothelial cells into the extracellular matrix in order to form new blood vessels contributing to the process of sprouting angiogenesis. |
| positive regulation of cell size | Any process that increases cell size. |
| positive regulation of endothelial cell proliferation | Any process that activates or increases the rate or extent of endothelial cell proliferation. |
| positive regulation of TOR signaling | Any process that activates or increases the frequency, rate or extent of TOR signaling. |
| positive regulation of vascular endothelial cell proliferation | Any process that activates or increases the frequency, rate or extent of vascular endothelial cell proliferation. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
32 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q01314 | AKT1 | RAC-alpha serine/threonine-protein kinase | Bos taurus (Bovine) | SS |
| Q8INB9 | Akt | RAC serine/threonine-protein kinase | Drosophila melanogaster (Fruit fly) | SS |
| O15530 | PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | Homo sapiens (Human) | EV |
| Q05513 | PRKCZ | Protein kinase C zeta type | Homo sapiens (Human) | SS |
| P41743 | PRKCI | Protein kinase C iota type | Homo sapiens (Human) | EV |
| Q16512 | PKN1 | Serine/threonine-protein kinase N1 | Homo sapiens (Human) | EV |
| Q6P5Z2 | PKN3 | Serine/threonine-protein kinase N3 | Homo sapiens (Human) | SS |
| Q16513 | PKN2 | Serine/threonine-protein kinase N2 | Homo sapiens (Human) | EV |
| P24723 | PRKCH | Protein kinase C eta type | Homo sapiens (Human) | SS |
| Q02156 | PRKCE | Protein kinase C epsilon type | Homo sapiens (Human) | SS |
| Q04759 | PRKCQ | Protein kinase C theta type | Homo sapiens (Human) | PR |
| Q05655 | PRKCD | Protein kinase C delta type | Homo sapiens (Human) | SS |
| P17252 | PRKCA | Protein kinase C alpha type | Homo sapiens (Human) | EV |
| P05129 | PRKCG | Protein kinase C gamma type | Homo sapiens (Human) | SS |
| P05771 | PRKCB | Protein kinase C beta type | Homo sapiens (Human) | SS |
| P31751 | AKT2 | RAC-beta serine/threonine-protein kinase | Homo sapiens (Human) | EV SS |
| P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| Q96BR1 | SGK3 | Serine/threonine-protein kinase Sgk3 | Homo sapiens (Human) | SS |
| Q9HBY8 | SGK2 | Serine/threonine-protein kinase Sgk2 | Homo sapiens (Human) | SS |
| O00141 | SGK1 | Serine/threonine-protein kinase Sgk1 | Homo sapiens (Human) | PR |
| Q15208 | STK38 | Serine/threonine-protein kinase 38 | Homo sapiens (Human) | EV |
| Q9Y2H1 | STK38L | Serine/threonine-protein kinase 38-like | Homo sapiens (Human) | EV |
| Q6A1A2 | PDPK2P | Putative 3-phosphoinositide-dependent protein kinase 2 | Homo sapiens (Human) | PR |
| P31750 | Akt1 | RAC-alpha serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q60823 | Akt2 | RAC-beta serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q9WUA6 | Akt3 | RAC-gamma serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| P47197 | Akt2 | RAC-beta serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| P47196 | Akt1 | RAC-alpha serine/threonine-protein kinase | Rattus norvegicus (Rat) | PR |
| Q63484 | Akt3 | RAC-gamma serine/threonine-protein kinase | Rattus norvegicus (Rat) | PR |
| Q9XTG7 | akt-2 | Serine/threonine-protein kinase akt-2 | Caenorhabditis elegans | SS |
| Q17941 | akt-1 | Serine/threonine-protein kinase akt-1 | Caenorhabditis elegans | PR |
| Q9SUA3 | D6PKL1 | Serine/threonine-protein kinase D6PKL1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSDVTIVKEG | WVQKRGEYIK | NWRPRYFLLK | TDGSFIGYKE | KPQDVDLPYP | LNNFSVAKCQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LMKTERPKPN | TFIIRCLQWT | TVIERTFHVD | TPEEREEWTE | AIQAVADRLQ | RQEEERMNCS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PTSQIDNIGE | EEMDASTTHH | KRKTMNDFDY | LKLLGKGTFG | KVILVREKAS | GKYYAMKILK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KEVIIAKDEV | AHTLTESRVL | KNTRHPFLTS | LKYSFQTKDR | LCFVMEYVNG | GELFFHLSRE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RVFSEDRTRF | YGAEIVSALD | YLHSGKIVYR | DLKLENLMLD | KDGHIKITDF | GLCKEGITDA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ATMKTFCGTP | EYLAPEVLED | NDYGRAVDWW | GLGVVMYEMM | CGRLPFYNQD | HEKLFELILM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EDIKFPRTLS | SDAKSLLSGL | LIKDPNKRLG | GGPDDAKEIM | RHSFFSGVNW | QDVYDKKLVP |
| 430 | 440 | 450 | 460 | 470 | |
| PFKPQVTSET | DTRYFDEEFT | AQTITITPPE | KYDEDGMDCM | DNERRPHFPQ | FSYSASGRE |