Q9XT26
Gene name |
CCNT1 |
Protein name |
Cyclin-T1 |
Names |
CycT1, Cyclin-T |
Species |
Equus caballus (Horse) |
KEGG Pathway |
ecb:100033893 |
EC number |
|
Protein Class |
|
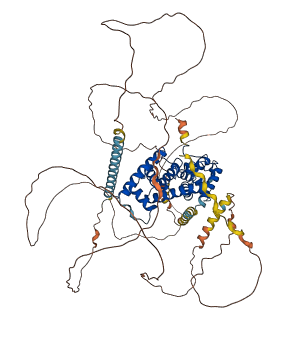
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
13-259 (N-terminal cyclin box) |
Relief mechanism |
PTM, Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q9XT26
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2W2H | X-ray | 325 A | A/B | 5-267 | PDB |
| AF-Q9XT26-F1 | Predicted | AlphaFoldDB |
No variants for Q9XT26
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q9XT26 | |||||
No associated diseases with Q9XT26
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cyclin/CDK positive transcription elongation factor complex | A transcription elongation factor complex that facilitates the transition from abortive to productive elongation by phosphorylating the CTD domain of the large subunit of DNA-directed RNA polymerase II, holoenzyme. Contains a cyclin and a cyclin-dependent protein kinase catalytic subunit. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| 7SK snRNA binding | Binding to a 7SK small nuclear RNA (7SK snRNA). |
| cyclin-dependent protein serine/threonine kinase activator activity | Binds to and increases the activity of a cyclin-dependent protein serine/threonine kinase. |
| cyclin-dependent protein serine/threonine kinase regulator activity | Modulates the activity of a cyclin-dependent protein serine/threonine kinase, enzymes of the protein kinase family that are regulated through association with cyclins and other proteins. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| positive regulation of DNA-templated transcription, elongation | Any process that activates or increases the frequency, rate or extent of transcription elongation, the extension of an RNA molecule after transcription initiation and promoter clearance by the addition of ribonucleotides catalyzed by a DNA-dependent RNA polymerase. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| transcription, DNA-templated | The synthesis of an RNA transcript from a DNA template. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q6T8E9 | CCNT1 | Cyclin-T1 | Bos taurus (Bovine) | SS |
| Q8HXN7 | CCNT1 | Cyclin-T1 | Pan troglodytes (Chimpanzee) | SS |
| O60563 | CCNT1 | Cyclin-T1 | Homo sapiens (Human) | EV |
| Q9QWV9 | Ccnt1 | Cyclin-T1 | Mus musculus (Mouse) | SS |
| Q2QQS5 | CYCT1-1 | Cyclin-T1-4 | Oryza sativa subsp japonica (Rice) | PR |
| Q6Z7H3 | CYCT1_2 | Cyclin-T1-2 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAC5 | CYCT1-3 | Cyclin-T1-3 | Oryza sativa subsp japonica (Rice) | PR |
| Q8GYM6 | CYCT1-4 | Cyclin-T1-4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEGERKNNNK | RWYFTREQLE | NSPSRRFGLD | PDKELSYRQQ | AANLLQDMGQ | RLNVSQLTIN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TAIVYMHRFY | MIQSFTQFHR | NSVAPAALFL | AAKVEEQPKK | LEHVIKVAHA | CLHPQESLPD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TRSEAYLQQV | QDLVILESII | LQTLGFELTI | DHPHTHVVKC | TQLVRASKDL | AQTSYFMATN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SLHLTTFSLQ | YTPPVVACVC | IHLACKWSNW | EIPVSTDGKH | WWEYVDATVT | LELLDELTHE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FLQILEKTPN | RLKRIRNWRA | CQAAKKTKAD | DRGTDENTSE | QTILNMISQS | SSDTTIAGLM |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SMSTSSTTST | VPSLPTTEES | SSNLSGVEML | QGERWLSSQP | PFKLEPAQGH | RTSENLALIG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VDHSLQQDGS | NAFISQKQNS | SKSVPSAKVS | LKEYRAKHAE | ELAAQKRQLE | NMEANVKSQY |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AYAAQNLLSH | HDSHSSVILK | MPIEGSENPE | RPFLEKPDKT | ALKMRIPVAS | GDKAASSKPE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| EIKMRIKVHA | APDKHNSIDD | SVTKSREHKE | KHKTHPSNHH | HHHNHHSHKH | SHSQLPAGTG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| NKRPGDPKHS | SQTSTLAHKT | YSLSSSFSSS | SSSRKRGPPE | ETGGALFDHP | AKIAKSTKSS |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SINFFPPLPT | MAQLPGHSSD | TSGLPFSQPS | CKTRVPHMKL | DKGPTGANGH | NTTQTIDYQD |
| 670 | 680 | 690 | 700 | 710 | 720 |
| TVNMLHSLLH | AQGVQPTQPP | ALEFVHSYGE | YLNPRAGGMP | SRSGNTDKPR | LPPLPSEPPP |
| PLPPLPK |