Q9WTZ2
Gene name |
Mbtps1 (S1p, Ski1) |
Protein name |
Membrane-bound transcription factor site-1 protease |
Names |
Endopeptidase S1P, Sterol-regulated luminal protease, Subtilisin/kexin isozyme 1, SKI-1 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:56453 |
EC number |
3.4.21.112: Serine endopeptidases |
Protein Class |
|
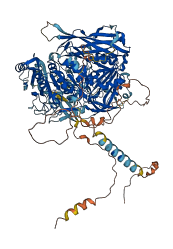
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9WTZ2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9WTZ2-F1 | Predicted | AlphaFoldDB |
40 variants for Q9WTZ2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3399757927 | 30 | L>F | No | EVA | |
| rs3399384452 | 31 | E>Q | No | EVA | |
| rs3389015507 | 32 | K>E | No | EVA | |
| rs3389020448 | 51 | V>SDG* | No | EVA | |
| rs3388984395 | 56 | Y>F | No | EVA | |
| rs3389008757 | 87 | I>V | No | EVA | |
| rs3388991243 | 153 | S>N | No | EVA | |
| rs3388965785 | 201 | V>M | No | EVA | |
| rs3388965707 | 241 | R>Q | No | EVA | |
| rs3389013282 | 286 | T>I | No | EVA | |
| rs3389017033 | 318 | F>S | No | EVA | |
| rs3389008737 | 345 | T>I | No | EVA | |
| rs3388991309 | 358 | V>L | No | EVA | |
| rs3389013498 | 422 | G>R | No | EVA | |
| rs3389015488 | 461 | Q>* | No | EVA | |
| rs3389007866 | 463 | H>D | No | EVA | |
| rs3388965726 | 463 | H>Q* | No | EVA | |
| rs3389020441 | 544 | N>I | No | EVA | |
| rs3389013691 | 554 | V>L | No | EVA | |
| rs3388984378 | 597 | S>N | No | EVA | |
| rs3388996859 | 610 | K>T | No | EVA | |
| rs3389007863 | 622 | R>G | No | EVA | |
| rs3389013633 | 647 | N>Y | No | EVA | |
| rs3399723000 | 762 | L>V | No | EVA | |
| rs3388984393 | 816 | K>E | No | EVA | |
| rs3388996845 | 840 | R>Q | No | EVA | |
| rs3389021991 | 860 | F>L | No | EVA | |
| rs3389017023 | 871 | S>T | No | EVA | |
| rs3412776890 | 895 | A>V | No | EVA | |
| rs3388996835 | 906 | H>R | No | EVA | |
| rs3389015489 | 911 | V>L | No | EVA | |
| rs3389013290 | 923 | P>L | No | EVA | |
| rs3389015661 | 951 | K>M | No | EVA | |
| rs3389013232 | 994 | Y>* | No | EVA | |
| rs3389013304 | 1030 | R>E* | No | EVA | |
| rs3389015463 | 1032 | R>* | No | EVA | |
| rs3389002519 | 1033 | P>A | No | EVA | |
| rs3389002519 | 1033 | P>S | No | EVA | |
| rs265745076 | 1047 | A>S | No | EVA | |
| rs3389017032 | 1049 | T>A | No | EVA |
No associated diseases with Q9WTZ2
4 regional properties for Q9WTZ2
Functions
| Description | ||
|---|---|---|
| EC Number | 3.4.21.112 | Serine endopeptidases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| Golgi stack | The set of thin, flattened membrane-bounded compartments, called cisternae, that form the central portion of the Golgi complex. The stack usually comprises cis, medial, and trans cisternae; the cis- and trans-Golgi networks are not considered part of the stack. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| metalloendopeptidase activity | Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain by a mechanism in which water acts as a nucleophile, one or two metal ions hold the water molecule in place, and charged amino acid side chains are ligands for the metal ions. |
| serine-type endopeptidase activity | Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine). |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| cholesterol metabolic process | The chemical reactions and pathways involving cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones. It is a component of the plasma membrane lipid bilayer and of plasma lipoproteins and can be found in all animal tissues. |
| lipid metabolic process | The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids. |
| lysosome organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a lysosome. A lysosome is a cytoplasmic, membrane-bounded organelle that is found in most animal cells and that contains a variety of hydrolases. |
| protein processing | Any protein maturation process achieved by the cleavage of a peptide bond or bonds within a protein. Protein maturation is the process leading to the attainment of the full functional capacity of a protein. |
| proteolysis | The hydrolysis of proteins into smaller polypeptides and/or amino acids by cleavage of their peptide bonds. |
| regulation of vesicle-mediated transport | Any process that modulates the rate, frequency, or extent of vesicle-mediated transport, the directed movement of substances, either within a vesicle or in the vesicle membrane, into, out of or within a cell. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q14703 | MBTPS1 | Membrane-bound transcription factor site-1 protease | Homo sapiens (Human) | PR |
| Q80W65 | Pcsk9 | Proprotein convertase subtilisin/kexin type 9 | Mus musculus (Mouse) | SS |
| Q9WTZ3 | Mbtps1 | Membrane-bound transcription factor site-1 protease | Rattus norvegicus (Rat) | PR |
| Q0WUG6 | SBT6.1 | Subtilisin-like protease SBT6.1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKLVSTWLLV | LVVLLCGKRH | LGDRLGTRAL | EKAPCPSCSH | LTLKVEFSST | VVEYEYIVAF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NGYFTAKARN | SFISSALKSS | EVENWRIIPR | NNPSSDYPSD | FEVIQIKEKQ | KAGLLTLEDH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PNIKRVTPQR | KVFRSLKFAE | SNPIVPCNET | RWSQKWQSSR | PLKRASLSLG | SGFWHATGRH |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SSRRLLRAIP | RQVAQTLQAD | VLWQMGYTGA | NVRVAVFDTG | LSEKHPHFKN | VKERTNWTNE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RTLDDGLGHG | TFVAGVIASM | RECQGFAPDA | ELHIFRVFTN | NQVSYTSWFL | DAFNYAILKK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MDVLNLSIGG | PDFMDHPFVD | KVWELTANNV | IMVSAIGNDG | PLYGTLNNPA | DQMDVIGVGG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| IDFEDNIARF | SSRGMTTWEL | PGGYGRVKPD | IVTYGAGVRG | SGVKGGCRAL | SGTSVASPVV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AGAVTLLVST | VQKRELVNPA | SVKQALIASA | RRLPGVNMFE | QGHGKLDLLR | AYQILSSYKP |
| 490 | 500 | 510 | 520 | 530 | 540 |
| QASLSPSYID | LTECPYMWPY | CSQPIYYGGM | PTIVNVTILN | GMGVTGRIVD | KPEWRPYLPQ |
| 550 | 560 | 570 | 580 | 590 | 600 |
| NGDNIEVAFS | YSSVLWPWSG | YLAISISVTK | KAASWEGIAQ | GHIMITVASP | AETELHSGAE |
| 610 | 620 | 630 | 640 | 650 | 660 |
| HTSTVKLPIK | VKIIPTPPRS | KRVLWDQYHN | LRYPPGYFPR | DNLRMKNDPL | DWNGDHVHTN |
| 670 | 680 | 690 | 700 | 710 | 720 |
| FRDMYQHLRS | MGYFVEVLGA | PFTCFDATQY | GTLLLVDSEE | EYFPEEIAKL | RRDVDNGLSL |
| 730 | 740 | 750 | 760 | 770 | 780 |
| VIFSDWYNTS | VMRKVKFYDE | NTRQWWMPDT | GGANIPALNE | LLSVWNMGFS | DGLYEGEFVL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| ANHDMYYASG | CSIAKFPEDG | VVITQTFKDQ | GLEVLKQETA | VVENVPILGL | YQIPSEGGGR |
| 850 | 860 | 870 | 880 | 890 | 900 |
| IVLYGDSNCL | DDSHRQKDCF | WLLDALLQYT | SYGVTPPSLS | HSGNRQRPPS | GAGLAPPERM |
| 910 | 920 | 930 | 940 | 950 | 960 |
| EGNHLHRYSK | VLEAHLGDPK | PRPLPACPHL | SWAKPQPLNE | TAPSNLWKHQ | KLLSIDLDKV |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| VLPNFRSNRP | QVRPLSPGES | GAWDIPGGIM | PGRYNQEVGQ | TIPVFAFLGA | MVALAFFVVQ |
| 1030 | 1040 | 1050 | |||
| ISKAKSRPKR | RRPRAKRPQL | AQQAHPARTP | SV |