Q9VXE5
Gene name |
mbt |
Protein name |
Serine/threonine-protein kinase PAK mbt |
Names |
Protein mushroom bodies tiny, p21-activated kinase-related protein |
Species |
Drosophila melanogaster (Fruit fly) |
KEGG Pathway |
dme:Dmel_CG18582 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
(Annotation based on sequence homology with Q9NQU5)
The p21-activated protein kinases5 (PAK5) belongs to group II PAK family and is involved in actin cytoskeleton regulation, cell growth, migration and survival, and so on. Group II PAKs (PAK4, 5, and 6) harbor well-conserved pseudosubstrate sequences. For PAK4, the N-terminal inhibitory segment contains two pseudopeptide sequences (FTGLPR at 32th and RPKPLV at 49th) that bind to the active site of the kinase domain as a pseudosubstrate. Like PAK4, PAK6 catalytic activity is inhibited by a peptide corresponding to its N-terminal pseudosubstrate (RRPKPVVDP at 49th).
PAKs are activated by the binding of GTP-loaded Cdc42 (or Rac) to the CRIB domain, which disrupts the dimer and unfolds autoinhibitory region. After releasing autoinhibitory region, kinase domain is then autophosphorylated for full activation.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
504-527 (Activation loop from InterPro)
Target domain |
368-619 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
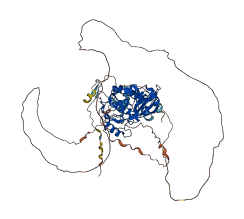
Autoinhibited structure

Activated structure

1 structures for Q9VXE5
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9VXE5-F1 | Predicted | AlphaFoldDB |
No variants for Q9VXE5
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q9VXE5 | |||||
No associated diseases with Q9VXE5
10 regional properties for Q9VXE5
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | Leucine-rich repeat | 594 - 652 | IPR001611-1 |
| repeat | Leucine-rich repeat | 664 - 708 | IPR001611-2 |
| repeat | Leucine-rich repeat | 710 - 766 | IPR001611-3 |
| repeat | Leucine-rich repeat, typical subtype | 614 - 637 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 639 - 661 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 662 - 685 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 687 - 706 | IPR003591-4 |
| repeat | Leucine-rich repeat, typical subtype | 708 - 731 | IPR003591-5 |
| repeat | Leucine-rich repeat, typical subtype | 754 - 777 | IPR003591-6 |
| domain | LRRC8, pannexin-like TM region | 1 - 341 | IPR021040 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| adherens junction | A cell-cell junction composed of the epithelial cadherin-catenin complex. The epithelial cadherins, or E-cadherins, of each interacting cell extend through the plasma membrane into the extracellular space and bind to each other. The E-cadherins bind to catenins on the cytoplasmic side of the membrane, where the E-cadherin-catenin complex binds to cytoskeletal components and regulatory and signaling molecules. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| small GTPase binding | Binding to a small monomeric GTPase. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| compound eye development | The process whose specific outcome is the progression of the compound eye over time, from its formation to the mature structure. The compound eye is an organ of sight that contains multiple repeating units, often arranged hexagonally. Each unit has its own lens and photoreceptor cell(s) and can generate either a single pixelated image or multiple images, per eye. |
| compound eye photoreceptor cell differentiation | The process in which a relatively unspecialized cell acquires the specialized features of an eye photoreceptor cell. |
| cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures. |
| Golgi organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the Golgi apparatus. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| mushroom body development | The process whose specific outcome is the progression of the mushroom body over time, from its formation to the mature structure. The mushroom body is composed of the prominent neuropil structures of the insect central brain, thought to be crucial for olfactory associated learning. These consist mainly of a bulbous calyx and tightly packaged arrays of thin parallel fibers of the Kenyon cells. |
| negative regulation of cell size | Any process that reduces cell size. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of compound eye photoreceptor development | Any process that activates or increases the frequency, rate or extent of compound eye photoreceptor development. |
| positive regulation of developmental growth | Any process that activates, maintains or increases the rate of developmental growth. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of cell-cell adhesion mediated by cadherin | Any process that modulates the frequency, rate or extent of cell-cell adhesion mediated by cadherin. |
| regulation of MAPK cascade | Any process that modulates the frequency, rate or extent of signal transduction mediated by the MAP kinase (MAPK) cascade. |
22 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q12469 | SKM1 | Serine/threonine-protein kinase SKM1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q08E52 | PAK1 | Serine/threonine-protein kinase PAK 1 | Bos taurus (Bovine) | SS |
| Q7YQL4 | PAK3 | Serine/threonine-protein kinase PAK 3 | Pan troglodytes (Chimpanzee) | SS |
| Q13153 | PAK1 | Serine/threonine-protein kinase PAK 1 | Homo sapiens (Human) | EV |
| Q13177 | PAK2 | Serine/threonine-protein kinase PAK 2 | Homo sapiens (Human) | EV |
| O75914 | PAK3 | Serine/threonine-protein kinase PAK 3 | Homo sapiens (Human) | SS |
| Q9P286 | PAK5 | Serine/threonine-protein kinase PAK 5 | Homo sapiens (Human) | EV |
| Q9NQU5 | PAK6 | Serine/threonine-protein kinase PAK 6 | Homo sapiens (Human) | EV |
| O96013 | PAK4 | Serine/threonine-protein kinase PAK 4 | Homo sapiens (Human) | EV |
| Q8CIN4 | Pak2 | Serine/threonine-protein kinase PAK 2 | Mus musculus (Mouse) | SS |
| O88643 | Pak1 | Serine/threonine-protein kinase PAK 1 | Mus musculus (Mouse) | SS |
| Q61036 | Pak3 | Serine/threonine-protein kinase PAK 3 | Mus musculus (Mouse) | SS |
| Q3ULB5 | Pak6 | Serine/threonine-protein kinase PAK 6 | Mus musculus (Mouse) | PR |
| Q8C015 | Pak5 | Serine/threonine-protein kinase PAK 5 | Mus musculus (Mouse) | SS |
| Q8BTW9 | Pak4 | Serine/threonine-protein kinase PAK 4 | Mus musculus (Mouse) | PR |
| Q62829 | Pak3 | Serine/threonine-protein kinase PAK 3 | Rattus norvegicus (Rat) | SS |
| Q64303 | Pak2 | Serine/threonine-protein kinase PAK 2 | Rattus norvegicus (Rat) | SS |
| P35465 | Pak1 | Serine/threonine-protein kinase PAK 1 | Rattus norvegicus (Rat) | SS |
| D4A280 | Pak5 | Serine/threonine-protein kinase PAK 5 | Rattus norvegicus (Rat) | SS |
| Q17850 | pak-1 | Serine/threonine-protein kinase pak-1 | Caenorhabditis elegans | PR |
| G5EGQ3 | max-2 | Serine/threonine-protein kinase max-2 | Caenorhabditis elegans | SS |
| G5EFU0 | pak-2 | Serine/threonine-protein kinase pak-2 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MFSKKKKKPL | ISMPSNFEHR | VHTGFDKREN | KYVGLPLQWA | SIVGNNQILK | SSNRPLPLVD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PSEITPTEIL | DLKTIVRPHH | NNNKADTTSL | NSSSTMMMGS | MAPMNPMAPG | AHPMMSHGPG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MMMPPETGGI | VLPKTSHVAR | SNSLRSSSPP | RVRRVANVPP | SVPEEEGPPA | AGTPGVGGAS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SGGFKPPGAH | PSLLYNSQHA | HANGATGPLA | VRTDQTNLQQ | YRSNLAPPSG | GSMPQQQQTS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PVGSVASGTR | SNHSHTNNGN | SGGSYPPMYP | TSHQQQQQQQ | QQAKQGGDQN | QNPLHPHAHP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HPHHHQHLAK | SASRASSSSG | GASSAAQQAS | GASGGAAGQP | KQDQRLTHEQ | FRAALQMVVS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| AGDPRENLDH | FNKIGEGSTG | TVCIATDKST | GRQVAVKKMD | LRKQQRRELL | FNEVVIMRDY |
| 430 | 440 | 450 | 460 | 470 | 480 |
| HHPNIVETYS | SFLVNDELWV | VMEYLEGGAL | TDIVTHSRMD | EEQIATVCKQ | CLKALAYLHS |
| 490 | 500 | 510 | 520 | 530 | 540 |
| QGVIHRDIKS | DSILLAADGR | VKLSDFGFCA | QVSQELPKRK | SLVGTPYWMS | PEVISRLPYG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| PEVDIWSLGI | MVIEMVDGEP | PFFNEPPLQA | MRRIRDMQPP | NLKNAHKVSP | RLQSFLDRML |
| 610 | 620 | 630 | |||
| VRDPAQRATA | AELLAHPFLR | QAGPPSLLVP | LMRNARHHP |