Q9VUC6
Gene name |
Frl |
Protein name |
Formin-like protein |
Names |
Formin related in leukocytes |
Species |
Drosophila melanogaster (Fruit fly) |
KEGG Pathway |
dme:Dmel_CG32138 |
EC number |
|
Protein Class |
FORMIN-LIKE PROTEIN (PTHR45857) |
Descriptions
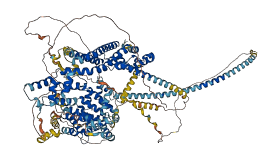
The diaphanous-related formin, Frl, the single fly member of the FMNL (formin related in leukocytes/formin-like) formin subfamily affects ommatidial rotation in the Drosophila eye and is controlled by the Rho family GTPase Cdc42. Full-length Frl is autoinhibited by the interaction between DAD domain and N-terminal region (probably DID domain as identified in other Flamins). Binding of Cdc42 to the GBD domain alleviates autoinhibition.
Autoinhibitory domains (AIDs)
Target domain |
687-1132 (FH2 domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment |
Target domain |
687-1132 (FH2 domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9VUC6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9VUC6-F1 | Predicted | AlphaFoldDB |
No variants for Q9VUC6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q9VUC6 | |||||
No associated diseases with Q9VUC6
5 regional properties for Q9VUC6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Formin, FH3 domain | 374 - 573 | IPR010472 |
| domain | Formin, GTPase-binding domain | 76 - 371 | IPR010473 |
| domain | Diaphanous autoregulatory (DAD) domain | 1136 - 1169 | IPR014767 |
| domain | Rho GTPase-binding/formin homology 3 (GBD/FH3) domain | 76 - 559 | IPR014768 |
| domain | Formin, FH2 domain | 687 - 1132 | IPR015425 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR45857 | FORMIN-LIKE PROTEIN |
| PANTHER Subfamily | PTHR45857:SF4 | FORMIN-LIKE PROTEIN |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| GTPase binding | Binding to a GTPase, any enzyme that catalyzes the hydrolysis of GTP. |
| small GTPase binding | Binding to a small monomeric GTPase. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| axon extension | Long distance growth of a single axon process involved in cellular development. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cortical actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of actin-based cytoskeletal structures in the cell cortex, i.e. just beneath the plasma membrane. |
| mushroom body development | The process whose specific outcome is the progression of the mushroom body over time, from its formation to the mature structure. The mushroom body is composed of the prominent neuropil structures of the insect central brain, thought to be crucial for olfactory associated learning. These consist mainly of a bulbous calyx and tightly packaged arrays of thin parallel fibers of the Kenyon cells. |
| ommatidial rotation | The process in which photoreceptors are arranged in ommatidia in the dorsal and ventral fields to be mirror images. The polarity is established in the imaginal discs concurrently with cell fate specification. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8IVF7 | FMNL3 | Formin-like protein 3 | Homo sapiens (Human) | SS |
| O95466 | FMNL1 | Formin-like protein 1 | Homo sapiens (Human) | SS |
| Q96PY5 | FMNL2 | Formin-like protein 2 | Homo sapiens (Human) | SS |
| Q6ZPF4 | Fmnl3 | Formin-like protein 3 | Mus musculus (Mouse) | SS |
| Q9JL26 | Fmnl1 | Formin-like protein 1 | Mus musculus (Mouse) | SS |
| A2APV2 | Fmnl2 | Formin-like protein 2 | Mus musculus (Mouse) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGAVKSRTIT | SADVDADEQL | QHPHSHAHHH | SMRNGHQHNG | SISSGTLQKQ | DLRYDIGCSS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QYQHVRQPSL | RSRSQQPMPT | TDELDRRFAK | VLASMDLPPD | KAKLLRNYDD | EKKWDMICDQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EMVQAKDPPS | HYLSKLRTYL | DPKASRSHRL | YLFYFLCQKR | KMVGESTSTQ | VLRDLEISLR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TNHIEWVKEF | LDDTNQGLDA | LVDYLSFRLQ | MMRHEQRLQG | VLCASEERLN | LTNGGDGGEI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VMGNSSSVSP | GGGGGLLSHG | NSTGHGLANG | TLDSRQQHTM | SYGFLRPTIA | DALDSPSLKR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RSRHIAKLNM | GAATDDIHVS | IMCLRAIMNN | KYGFNMVIQH | REAINCIALS | LIHKSLRTKA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LVLELLAAIC | LVKGGHEIIL | GSFDNFKDVC | QEKRRFQTLM | EYFMNFEAFN | IDFMVACMQF |
| 430 | 440 | 450 | 460 | 470 | 480 |
| MNIVVHSVED | MNYRVHLQYE | FTALGLDKYL | ERIRLTESEE | LKVQISAYLD | NVFDVAALME |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DSETKTSALE | RVQELEDQLE | REIDRNSEFL | YKYAELESES | LTLKTEREQL | AMIRQKLEEE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LTVMQRMLQH | NEQELKKRDT | LLHTKNMELQ | TLSRSLPRSA | SSGDGSLANG | GLMAGSTSGA |
| 610 | 620 | 630 | 640 | 650 | 660 |
| ASLTLPPPPP | PMPASPTASS | AAPPPPPPPA | PPAPPPPPGF | SPLGSPSGSL | ASTAPSPPHA |
| 670 | 680 | 690 | 700 | 710 | 720 |
| PPMLSSFQPP | PPPVAGFMPA | PDGAMTIKRK | VPTKYKLPTL | NWIALKPNQV | RGTIFNELDD |
| 730 | 740 | 750 | 760 | 770 | 780 |
| EKIFKQIDFN | EFEERFKIGI | GGALRNGSNG | TEVDGSLQSS | KRFKRPDNVS | LLEHTRLRNI |
| 790 | 800 | 810 | 820 | 830 | 840 |
| AISRRKLGMP | IDDVIAAIHS | LDLKKLSLEN | VELLQKMVPT | DAEVKSYKEY | IIERKDQQLL |
| 850 | 860 | 870 | 880 | 890 | 900 |
| TEEDKFMLQL | SRVERISSKL | AIMNYMGNFV | DSVHLISPQV | QSIAGASTSL | KQSRKFKAVL |
| 910 | 920 | 930 | 940 | 950 | 960 |
| EIVLAFGNYL | NSNKRGPAYG | FKLQSLDTLI | DTKSTDKRSS | LLHYIVATIR | AKFPELLNFE |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| SELYGTDKAA | SVALENVVAD | VQELEKGMDL | VRKEAELRVK | GAQTHILRDF | LNNSEDKLKK |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| IKSDLRHAQE | AFKECVEYFG | DSSRNADAAA | FFALIVRFTR | AFKQHDQENE | QRLRLEKAAA |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| LAASKKENDQ | VLMRNKVNQK | KQQLPSNGAL | SLQEAVINEL | KSKAHSVREK | KLLQQDEVYN |
| 1150 | 1160 | 1170 | 1180 | ||
| GALEDILLGL | KSEPYRRADA | VRRSQRRRID | NNRLSRTLEE | MDC |