Q9VE61
Gene name |
CG7694 |
Protein name |
E3 ubiquitin-protein ligase RNF181 homolog |
Names |
RING finger protein 181 homolog, RING-type E3 ubiquitin transferase RNF181 homolog |
Species |
Drosophila melanogaster (Fruit fly) |
KEGG Pathway |
dme:Dmel_CG7694 |
EC number |
2.3.2.27: Aminoacyltransferases |
Protein Class |
|
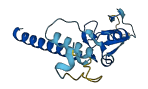
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9VE61
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9VE61-F1 | Predicted | AlphaFoldDB |
No variants for Q9VE61
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q9VE61 | |||||
No associated diseases with Q9VE61
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.27 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
| zinc ion binding | Binding to a zinc ion (Zn). |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| protein autoubiquitination | The ubiquitination by a protein of one or more of its own amino acid residues, or residues on an identical protein. Ubiquitination occurs on the lysine residue by formation of an isopeptide crosslink. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
17 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O00237 | RNF103 | E3 ubiquitin-protein ligase RNF103 | Homo sapiens (Human) | PR |
| Q86Y13 | DZIP3 | E3 ubiquitin-protein ligase DZIP3 | Homo sapiens (Human) | PR |
| Q9Y3E7 | CHMP3 | Charged multivesicular body protein 3 | Homo sapiens (Human) | EV |
| Q9R1W3 | Rnf103 | E3 ubiquitin-protein ligase RNF103 | Mus musculus (Mouse) | PR |
| Q9EPZ8 | Rnf103 | E3 ubiquitin-protein ligase RNF103 | Rattus norvegicus (Rat) | PR |
| Q9SI09 | XERICO | Probable E3 ubiquitin-protein ligase XERICO | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZV51 | ATL56 | RING-H2 finger protein ATL56 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SKK8 | ATL22 | RING-H2 finger protein ATL22 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0CH02 | ATL21B | Putative RING-H2 finger protein ATL21B | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0CH01 | ATL21A | Putative RING-H2 finger protein ATL21A | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22255 | ATL64 | RING-H2 finger protein ATL64 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RXX9 | ATL6 | E3 ubiquitin-protein ligase ATL6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8GYT9 | SIS3 | E3 ubiquitin-protein ligase SIS3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LZJ6 | ATL5 | RING-H2 finger protein ATL5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8GXF8 | SGR9 | E3 ubiquitin-protein ligase SGR9, amyloplastic | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LGA5 | ATL31 | E3 ubiquitin-protein ligase ATL31 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LUZ9 | ATL63 | RING-H2 finger protein ATL63 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSDYFEELGH | EPTGPLGAND | LARNLKRLQV | LAIMNGIDME | IEVPEASKRA | ILELPVHEIV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KSDEGGDLEC | SVCKEPAEEG | QKYRILPCKH | EFHEECILLW | LKKTNSCPLC | RYELETDDPV |
| 130 | 140 | ||||
| YEELRRFRQD | EANRRERENT | LLDSMFG |