Q9V461
Gene name |
Mcm6 |
Protein name |
DNA replication licensing factor Mcm6 |
Names |
DmMCM6 |
Species |
Drosophila melanogaster (Fruit fly) |
KEGG Pathway |
dme:Dmel_CG4039 |
EC number |
3.6.4.12: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
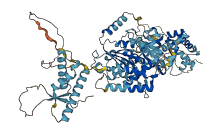
Autoinhibited structure

Activated structure

5 structures for Q9V461
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6RAW | EM | 370 A | 6 | 1-817 | PDB |
| 6RAX | EM | 399 A | 6 | 1-817 | PDB |
| 6RAY | EM | 428 A | 6 | 1-817 | PDB |
| 6RAZ | EM | 446 A | 6 | 1-817 | PDB |
| AF-Q9V461-F1 | Predicted | AlphaFoldDB |
No variants for Q9V461
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q9V461 | |||||
No associated diseases with Q9V461
6 regional properties for Q9V461
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | MCM domain | 326 - 548 | IPR001208 |
| conserved_site | Mini-chromosome maintenance, conserved site | 447 - 455 | IPR018525 |
| domain | MCM N-terminal domain | 22 - 95 | IPR027925 |
| domain | MCM OB domain | 116 - 245 | IPR033762 |
| domain | Mcm6, C-terminal winged-helix domain | 702 - 816 | IPR041024 |
| domain | MCM, AAA-lid domain | 564 - 648 | IPR041562 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.12 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| CMG complex | A protein complex that contains the GINS complex, Cdc45p, and the heterohexameric MCM complex, and that is involved in unwinding DNA during replication. |
| MCM complex | A hexameric protein complex required for the initiation and regulation of DNA replication. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| DNA helicase activity | Unwinding of a DNA helix, driven by ATP hydrolysis. |
| metal ion binding | Binding to a metal ion. |
| single-stranded DNA binding | Binding to single-stranded DNA. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| DNA replication | The cellular metabolic process in which a cell duplicates one or more molecules of DNA. DNA replication begins when specific sequences, known as origins of replication, are recognized and bound by initiation proteins, and ends when the original DNA molecule has been completely duplicated and the copies topologically separated. The unit of replication usually corresponds to the genome of the cell, an organelle, or a virus. The template for replication can either be an existing DNA molecule or RNA. |
| DNA replication initiation | The process in which DNA-dependent DNA replication is started; this begins with the ATP dependent loading of an initiator complex onto the DNA, this is followed by DNA melting and helicase activity. In bacteria, the gene products that enable the helicase activity are loaded after the initial melting and in archaea and eukaryotes, the gene products that enable the helicase activity are inactive when they are loaded and subsequently activate. |
| DNA unwinding involved in DNA replication | The process in which interchain hydrogen bonds between two strands of DNA are broken or 'melted', generating unpaired template strands for DNA replication. |
| double-strand break repair via break-induced replication | The error-free repair of a double-strand break in DNA in which the centromere-proximal end of a broken chromosome searches for a homologous region in an intact chromosome. DNA synthesis initiates from the 3' end of the invading DNA strand, using the intact chromosome as the template, and progresses to the end of the chromosome. |
| eggshell chorion gene amplification | Amplification by up to 60-fold of the loci containing the chorion gene clusters. Amplification is necessary for the rapid synthesis of chorion proteins by the follicle cells, and occurs by repeated firing of one or more origins located within each gene cluster. |
| mitotic DNA replication | Any nuclear DNA replication that is involved in a mitotic cell cycle. |
| regulation of DNA-templated DNA replication initiation | Any process that modulates the frequency, rate or extent of initiation of DNA-dependent DNA replication; the process in which DNA becomes competent to replicate. In eukaryotes, replication competence is established in early G1 and lost during the ensuing S phase. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KIZ8 | MCM6 | DNA replication licensing factor MCM6 | Bos taurus (Bovine) | PR |
| Q9XYU0 | Mcm7 | DNA replication licensing factor Mcm7 | Drosophila melanogaster (Fruit fly) | PR |
| Q14566 | MCM6 | DNA replication licensing factor MCM6 | Homo sapiens (Human) | PR |
| P97311 | Mcm6 | DNA replication licensing factor MCM6 | Mus musculus (Mouse) | PR |
| Q62724 | Mcm6 | DNA replication licensing factor MCM6 | Rattus norvegicus (Rat) | PR |
| Q6F353 | Os05g0235800 | DNA replication licensing factor MCM6 | Oryza sativa subsp japonica (Rice) | PR |
| P34647 | mcm-6 | DNA replication licensing factor mcm-6 | Caenorhabditis elegans | PR |
| Q9SF37 | MCM8 | Probable DNA helicase MCM8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q28CM3 | mmcm6 | Maternal DNA replication licensing factor mcm6 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q6P1V8 | zmcm6 | Zygotic DNA replication licensing factor mcm6 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDVADAQVGQ | LRVKDEVGIR | AQKLFQDFLE | EFKEDGEIKY | TRPAASLESP | DRCTLEVSFE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DVEKYDQNLA | TAIIEEYYHI | YPFLCQSVSN | YVKDRIGLKT | QKDCYVAFTE | VPTRHKVRDL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TTSKIGTLIR | ISGQVVRTHP | VHPELVSGVF | MCLDCQTEIR | NVEQQFKFTN | PTICRNPVCS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NRKRFMLDVE | KSLFLDFQKI | RIQETQAELP | RGCIPRAVEI | ILRSELVETV | QAGDRYDFTG |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TLIVVPDVSV | LAGVGTRAEN | SSRHKPGEGM | DGVTGLKALG | MRELNYRMAF | LACSVQATTA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RFGGTDLPMS | EVTAEDMKKQ | MTDAEWHKIY | EMSKDRNLYQ | NLISSLFPSI | YGNDEVKRGI |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LLQQFGGVAK | TTTEKTSLRG | DINVCIVGDP | STAKSQFLKQ | VSDFSPRAIY | TSGKASSAAG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LTAAVVRDEE | SFDFVIEAGA | LMLADNGICC | IDEFDKMDQR | DQVAIHEAME | QQTISIARAG |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VRATLNARTS | ILAAANPING | RYDRSKSLQQ | NIQLSAPIMS | RFDLFFILVD | ECNEVVDYAI |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ARKIVDLHSN | IEESVERAYT | REEVLRYVTF | ARQFKPVISQ | EAGHMLVENY | GHLRQRDTGT |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SGRSTWRITV | RQLESMIRLS | EAMAKLECSN | RVLERHVKEA | FRLLNKSIIR | VEQPDIHLDD |
| 670 | 680 | 690 | 700 | 710 | 720 |
| DEGLDMDDGI | QHDIDMENNG | AAANVDENNG | MDTSASGAVQ | KKKFTLSFED | YKNLSTMLVL |
| 730 | 740 | 750 | 760 | 770 | 780 |
| HMRAEEARCE | VEGNDTGIKR | SNVVTWYLEQ | VADQIESEDE | LISRKNLIEK | LIDRLIYHDQ |
| 790 | 800 | 810 | |||
| VIIPLKTSTL | KPRIQVQKDF | VEEDDPLLVV | HPNYVVE |