Q9UQM7
Gene name |
CAMK2A (CAMKA, KIAA0968) |
Protein name |
Calcium/calmodulin-dependent protein kinase type II subunit alpha |
Names |
CaM kinase II subunit alpha, CaMK-II subunit alpha |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:815 |
EC number |
2.7.11.17: Protein-serine/threonine kinases |
Protein Class |
SERINE/THREONINE-PROTEIN KINASE (PTHR24347) |
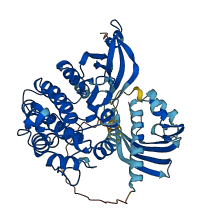
Descriptions
Calcium/calmodulin-dependent protein kinase II (CaMKII) is a serine/threonine kinase that is highly abundant in the brain, especially in the postsynaptic density (PSD). CaMKII forms a homomeric or heteromeric dodecamer of four different isoforms (α, β, γ, δ) encoded by four genes (CAMK2A, CAMK2B, CAMK2G, and CAMK2D, respectively), and α and β isoforms are predominant in the brain. In the resting state, the kinase activity of each subunit of CaMKII is inhibited by binding of the regulatory segment of CaMKII to the substrate binding groove of its kinase domain, which is called autoinhibition. When Ca2+-bound calmodulin (Ca2+/CaM) increases upon intracellular Ca2+ elevation, Ca2+/CaM binding to the regulatory segment dissociates the segment from the kinase domain, and within the regulatory segment, Thr286 in CaMKIIα and Thr287 in CaMKIIβ are phosphorylated by adjacent subunits, preventing the segment from returning to the kinase domain. This CaMKII activity is essential for neuronal plasticity and learning.
Autoinhibitory domains (AIDs)
Target domain |
13-271 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
Structural analysis, Mutagenesis experiment |
Accessory elements
155-178 (Activation loop from InterPro)
Target domain |
13-271 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Akita T et al. (2018) "De novo variants in CAMK2A and CAMK2B cause neurodevelopmental disorders", Annals of clinical and translational neurology, 5, 280-296
- Yang E et al. (1999) "Structural examination of autoregulation of multifunctional calcium/calmodulin-dependent protein kinase II", The Journal of biological chemistry, 274, 26199-208
- Rellos P et al. (2010) "Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation", PLoS biology, 8, e1000426
- Özden C et al. (2022) "CaMKII binds both substrates and activators at the active site", Cell reports, 40, 111064
Autoinhibited structure

Activated structure

22 structures for Q9UQM7
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2VZ6 | X-ray | 230 A | A/B | 13-302 | PDB |
| 3SOA | X-ray | 355 A | A | 1-474 | PDB |
| 5IG3 | X-ray | 275 A | A/B/C/D/E/F | 345-475 | PDB |
| 6OF8 | X-ray | 210 A | A/B/C/D/E/F/G | 345-475 | PDB |
| 6VZK | X-ray | 255 A | A | 7-274 | PDB |
| 6W4O | EM | 480 A | A/B/C/D/E/F/G/I/J/K/L/M/O | 7-478 | PDB |
| 6W4P | EM | 660 A | A/B/C/D/E/F/G/H/I/J/K/L/M | 7-478 | PDB |
| 6X5G | X-ray | 185 A | A | 7-274 | PDB |
| 6X5Q | X-ray | 214 A | A | 7-274 | PDB |
| 7KL0 | X-ray | 240 A | A/B | 7-274 | PDB |
| 7KL1 | X-ray | 240 A | A/B | 7-274 | PDB |
| 7KL2 | X-ray | 256 A | A | 7-274 | PDB |
| 7REC | X-ray | 220 A | A/B/C/D/E/F/G | 345-475 | PDB |
| 7UIQ | X-ray | 311 A | A/B | 7-274 | PDB |
| 7UIR | X-ray | 310 A | A/B | 7-274 | PDB |
| 7UIS | X-ray | 258 A | A | 7-274 | PDB |
| 7UJP | X-ray | 256 A | A/B | 7-274 | PDB |
| 7UJQ | X-ray | 225 A | A/B | 7-274 | PDB |
| 7UJR | X-ray | 195 A | A | 7-274 | PDB |
| 7UJS | X-ray | 275 A | A | 7-274 | PDB |
| 7UJT | X-ray | 210 A | A | 7-274 | PDB |
| AF-Q9UQM7-F1 | Predicted | AlphaFoldDB |
178 variants for Q9UQM7
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1554122526 RCV000577897 CA361754600 VAR_080579 RCV000516156 |
98 | F>S | Intellectual disability Intellectual disability, autosomal dominant 53 MRD53; no effect on protein abundance; decreased autophosphorylation; decreased neuronal migration [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1287121256 RCV000577915 CA361754204 RCV000516151 VAR_080580 |
109 | E>D | Intellectual disability Intellectual disability, autosomal dominant 53 MRD53; no effect on protein abundance; increased autophosphorylation; decreased neuronal migration [ClinVar, UniProt] | Yes |
ClinGen ClinVar dbSNP gnomAD UniProt |
|
rs779607303 CA3509897 RCV001251933 |
110 | A>V | Variant assessed as Somatic; 0.0 impact. Intellectual disability, autosomal dominant 53 [NCI-TCGA, ClinVar] | Yes |
ClinGen ClinVar ExAC NCI-TCGA dbSNP gnomAD |
| VAR_080581 | 112 | A>V | MRD53; unknown pathological significance [UniProt] | Yes | UniProt |
| VAR_080582 | 138 | P>A | MRD53; unknown pathological significance; no effect on protein abundance; no effect on autophosphorylation; no effect on neuronal migration [UniProt] | Yes | UniProt |
|
CA361750749 RCV000577883 RCV000516153 rs1554122129 VAR_080583 |
183 | E>V | Intellectual disability Intellectual disability, autosomal dominant 53 MRD53; increased ubiquitin-mediated proteasomal degradation with a dominant negative effect on wild-type protein; decreased localization to dendritic spines; no effect on holoenzyme assembly; loss of interaction with SHANK3; loss of interaction with GRIN2B; loss of interaction with CACNB2; loss of interaction with LRRC7; loss of interaction with GRM5; decreased protein serine/threonine kinase activity with a dominant negative effect on wild-type protein; decreased autophosphorylation; changed dendritic spine development; decreased neuronal migration [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA129150428 RCV001558478 rs926027867 RCV002470885 VAR_080584 RCV000577918 |
212 | P>L | Intellectual disability Variant assessed as Somatic; impact. Intellectual disability, autosomal dominant 53 MRD53; unknown pathological significance; no effect on protein abundance; no effect on autophosphorylation; no effect on neuronal migration [ClinVar, NCI-TCGA, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl NCI-TCGA dbSNP |
|
rs926027867 VAR_081160 RCV000678211 CA361750093 |
212 | P>Q | Intellectual disability, autosomal dominant 53 MRD53; increased basal autophosphorylation [ClinVar, UniProt] | Yes |
ClinGen ClinVar Ensembl dbSNP UniProt |
|
CA248921 RCV000577881 RCV000678212 rs864309606 VAR_080585 RCV000202726 |
235 | P>L | Intellectual disability Intellectual disability, autosomal dominant 53 MRD53; unknown pathological significance; no effect on protein abundance; no effect on autophosphorylation; no effect on neuronal migration [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1554121875 CA361749404 VAR_080586 RCV000516155 RCV000577902 |
282 | H>R | Intellectual disability Intellectual disability, autosomal dominant 53 MRD53; decreased protein abundance; increased autophosphorylation; decreased neuronal migration [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000516152 CA361749377 rs1554121872 RCV000577925 VAR_080587 |
286 | T>P | Intellectual disability Intellectual disability, autosomal dominant 53 MRD53; no effect on protein abundance; loss of autophosphorylation; loss of neuronal migration [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1754406562 RCV001266835 |
444 | P>T | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
CA361739033 VAR_081161 RCV000678210 rs1554119274 |
466 | H>Y | Intellectual disability, autosomal recessive 63 MRT63; decreased oligomerization [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA3509993 rs757273062 |
2 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1373405268 CA361723264 |
3 | T>I | No |
ClinGen gnomAD |
|
|
rs1056874482 CA129116861 |
8 | R>C | No |
ClinGen TOPMed |
|
|
CA3509991 rs764230609 |
8 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs6861622 CA361723117 |
10 | T>A | No |
ClinGen Ensembl |
|
|
rs1160900025 CA361723102 |
10 | T>M | No |
ClinGen TOPMed |
|
|
rs6861622 CA361723114 |
10 | T>S | No |
ClinGen Ensembl |
|
|
CA129116852 rs928636452 |
12 | E>D | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 14 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1064323 rs1580966945 CA361722927 |
17 | E>K | endometrium [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs1202019049 CA361718950 |
23 | A>G | No |
ClinGen gnomAD |
|
|
rs1439139668 CA361718935 COSM298063 |
25 | S>L | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs751885208 CA3509969 |
28 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA361718920 rs1331999283 |
28 | R>Q | No |
ClinGen gnomAD |
|
|
rs766825548 CA3509968 |
38 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA361718825 rs1580947248 |
39 | Y>D | No |
ClinGen Ensembl |
|
|
rs1267862122 CA361718776 |
43 | I>V | No |
ClinGen TOPMed |
|
|
rs1356458925 CA361718731 |
46 | T>S | No |
ClinGen gnomAD |
|
| TCGA novel | 47 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs202217757 CA129109536 |
52 | R>T | No |
ClinGen Ensembl |
|
|
rs750935357 CA3509948 |
54 | H>L | No |
ClinGen ExAC gnomAD |
|
|
rs761424198 CA3509946 |
59 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA3509943 rs544705728 |
62 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA129106222 rs867820640 |
62 | R>H | No |
ClinGen gnomAD |
|
| TCGA novel | 64 | C>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA361715968 rs1195589386 |
65 | R>H | No |
ClinGen TOPMed |
|
|
CA129102100 rs61732056 |
74 | R>* | No |
ClinGen Ensembl |
|
|
rs61732056 CA129102106 |
74 | R>G | No |
ClinGen Ensembl |
|
|
CA361714523 rs1366605186 |
75 | L>Q | No |
ClinGen TOPMed |
|
|
rs202107716 CA3509921 |
79 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA361714436 rs1201263122 |
81 | E>* | No |
ClinGen gnomAD |
|
|
rs1480404414 CA361714400 |
83 | G>E | No |
ClinGen gnomAD |
|
|
rs866698307 CA129102081 |
87 | L>P | No |
ClinGen Ensembl |
|
| TCGA novel | 98 | F>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 106 | Y>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 121 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA129154210 rs758821296 |
124 | L>V | No |
ClinGen Ensembl |
|
|
CA3509871 rs780817639 |
134 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA129151677 rs866152886 |
150 | A>V | No |
ClinGen Ensembl |
|
|
CA129151643 rs986491632 |
151 | A>T | No |
ClinGen Ensembl |
|
|
rs745482187 CA3509849 |
163 | V>L | No |
ClinGen ExAC |
|
|
rs1372333506 CA361752268 |
164 | E>D | No |
ClinGen gnomAD |
|
|
rs1755922883 RCV001200176 |
168 | Q>* | No |
ClinVar dbSNP |
|
|
CA3509848 rs778835439 |
169 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA361752117 rs1422349611 |
169 | A>V | No |
ClinGen TOPMed |
|
| TCGA novel | 170 | W>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs756158214 CA3509826 |
191 | G>W | No |
ClinGen ExAC gnomAD |
|
|
rs370600720 CA3509824 |
193 | P>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 195 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1462812363 CA361750187 |
207 | L>P | No |
ClinGen TOPMed |
|
|
rs747987338 CA3509790 |
210 | Y>S | No |
ClinGen ExAC gnomAD |
|
|
rs1300146469 CA361750002 |
217 | D>E | No |
ClinGen TOPMed |
|
|
CA129150416 rs11956781 |
219 | H>L | No |
ClinGen Ensembl |
|
|
CA361749979 rs11956781 |
219 | H>P | No |
ClinGen Ensembl |
|
|
rs1185191650 CA361749966 |
220 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1425310941 COSM1064319 CA361749963 |
220 | R>H | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA361749957 rs1425310941 |
220 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
rs17854002 CA361749912 |
223 | Q>* | No |
ClinGen gnomAD |
|
|
CA129150376 rs17854002 |
223 | Q>K | No |
ClinGen gnomAD |
|
|
CA361749806 rs1236371050 |
228 | G>S | No |
ClinGen gnomAD |
|
|
rs1319168744 CA361749795 |
229 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA3509782 rs368023327 |
231 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM1064318 CA361749737 rs1160671001 |
234 | S>L | Variant assessed as Somatic; 4.641e-05 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1325334771 CA361749714 |
238 | D>N | No |
ClinGen gnomAD |
|
|
CA3509761 rs751800259 |
242 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 255 | N>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA361749584 rs1580922542 |
257 | S>P | No |
ClinGen Ensembl |
|
|
rs915686916 CA129149561 |
260 | I>L | No |
ClinGen Ensembl |
|
|
rs1487504062 CA361749559 |
260 | I>M | No |
ClinGen gnomAD |
|
|
CA3509759 rs758925573 |
261 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA361749541 rs1346541754 |
264 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA3509757 rs764807945 |
265 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs568397508 CA129149541 |
268 | H>R | No |
ClinGen Ensembl |
|
|
CA361749502 rs1331653509 |
269 | P>L | No |
ClinGen gnomAD |
|
|
CA361749498 RCV000760641 rs1562164380 |
270 | W>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA3509737 rs200216623 |
274 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA361749398 rs1171881562 |
283 | R>K | No |
ClinGen gnomAD |
|
|
RCV001090380 rs1755639151 |
301 | G>E | No |
ClinVar dbSNP |
|
|
rs1210191385 CA361748310 |
303 | I>V | No |
ClinGen gnomAD |
|
|
CA361748244 rs1349742423 |
306 | T>M | No |
ClinGen gnomAD |
|
|
CA361748138 rs1449666180 |
315 | G>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1158829901 CA361747682 |
316 | G>R | No |
ClinGen TOPMed |
|
|
CA3509693 rs765165346 |
319 | G>E | No |
ClinGen ExAC gnomAD |
|
|
rs55696902 CA3509691 |
321 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 324 | S>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA361747498 rs375813680 |
324 | S>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1445974036 CA361747492 |
325 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1292054661 CA361745701 |
331 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs770841322 CA129139929 |
332 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA361745636 rs1350890212 |
334 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
CA361745632 rs1287488367 |
334 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1279296830 CA361745427 |
342 | D>E | No |
ClinGen TOPMed |
|
|
CA129139920 rs1024265518 |
343 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
CA129139917 rs999120588 |
343 | T>N | No |
ClinGen TOPMed |
|
|
rs1345838148 CA361745385 |
345 | V>L | No |
ClinGen gnomAD |
|
|
CA3509582 rs778942662 |
346 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs745982580 CA3509583 |
346 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 349 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 351 | I>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3509581 rs757512233 |
351 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA361742778 rs1190483281 |
353 | V>L | No |
ClinGen gnomAD |
|
|
CA361742681 rs1440684133 |
357 | L>M | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 359 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 360 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA361742511 rs1201903749 |
364 | G>E | No |
ClinGen gnomAD |
|
| TCGA novel | 365 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1301785267 CA361741197 |
376 | G>D | No |
ClinGen gnomAD |
|
|
rs1408224855 CA361741119 |
382 | P>S | No |
ClinGen gnomAD |
|
|
rs1408224855 CA361741120 |
382 | P>T | No |
ClinGen gnomAD |
|
|
rs1313874797 CA361741112 |
383 | E>Q | No |
ClinGen TOPMed |
|
|
rs764964453 CA3509553 |
387 | N>I | No |
ClinGen ExAC gnomAD |
|
|
CA3509552 rs761351468 |
393 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs1420436174 CA361740966 |
395 | H>R | No |
ClinGen gnomAD |
|
|
rs753708011 CA3509551 |
396 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA361740955 rs1442791113 |
396 | R>Q | No |
ClinGen gnomAD |
|
|
CA361740931 rs1204852152 |
398 | Y>C | No |
ClinGen gnomAD |
|
|
rs763794315 CA3509550 |
401 | N>T | No |
ClinGen ExAC gnomAD |
|
|
CA3509549 rs376699738 |
402 | L>M | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 403 | W>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA361740194 rs1161905638 |
405 | R>Q | No |
ClinGen gnomAD |
|
|
rs1362570011 CA361740198 |
405 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA3509519 rs772746827 |
410 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1580891745 CA361740118 |
412 | T>P | No |
ClinGen Ensembl |
|
|
CA361740112 rs1580891738 |
413 | T>P | No |
ClinGen Ensembl |
|
|
rs1269084885 CA361740071 |
417 | P>T | No |
ClinGen gnomAD |
|
|
rs756733402 CA3509516 |
419 | I>F | No |
ClinGen ExAC gnomAD |
|
|
CA361740015 rs1580891693 |
420 | H>P | No |
ClinGen Ensembl |
|
|
rs1311432465 CA361739969 |
422 | M>I | No |
ClinGen gnomAD |
|
|
CA361739947 rs1406491152 |
424 | D>N | No |
ClinGen gnomAD |
|
|
RCV001310532 rs1754415187 |
425 | E>K | No |
ClinVar dbSNP |
|
|
rs907583897 CA129123696 |
429 | I>V | No |
ClinGen Ensembl |
|
|
rs752541567 CA3509512 |
430 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA361739819 rs1266065955 |
432 | I>V | No |
ClinGen Ensembl |
|
|
CA3509511 rs767291853 |
433 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754962340 CA3509510 |
433 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs766270888 CA3509508 |
435 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs760949012 CA3509504 |
440 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA361739651 rs374924411 |
440 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs374924411 CA3509505 |
440 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA129123595 rs199592588 |
441 | G>S | No |
ClinGen 1000Genomes |
|
|
rs772552968 CA3509502 |
442 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA361739558 rs1221317581 |
445 | R>H | No |
ClinGen TOPMed |
|
|
CA361739552 rs1456960358 |
446 | T>A | No |
ClinGen TOPMed |
|
| TCGA novel | 446 | T>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA361739551 rs1456960358 |
446 | T>P | No |
ClinGen TOPMed |
|
|
CA3509498 rs748774194 |
447 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA361739520 rs1580891529 |
447 | A>V | No |
ClinGen Ensembl |
|
|
rs777247730 CA3509497 |
449 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA361739435 rs1580891510 |
452 | T>P | No |
ClinGen Ensembl |
|
|
rs367551385 CA3509495 |
453 | R>C | No |
ClinGen ESP ExAC gnomAD |
|
|
CA3509494 rs780762936 |
453 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA361739407 rs1382795728 |
454 | V>F | No |
ClinGen gnomAD |
|
|
CA361739412 rs1382795728 |
454 | V>I | No |
ClinGen gnomAD |
|
|
rs1580891481 CA361739370 |
456 | H>P | No |
ClinGen Ensembl |
|
|
CA3509493 rs754732557 |
457 | R>C | Variant assessed as Somatic; 4.647e-05 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs375918851 CA3509492 |
457 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs577586528 CA361739324 |
458 | R>W | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1167006357 CA361739218 |
460 | G>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA3509489 rs753904024 |
464 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764264581 CA3509488 |
465 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA361738952 rs954139326 |
468 | H>Q | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 472 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3509485 rs767783317 |
472 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs771394411 CA3509482 |
475 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA3509481 rs763490198 |
477 | P>H | No |
ClinGen ExAC gnomAD |
2 associated diseases with Q9UQM7
[MIM: 617798]: Intellectual developmental disorder, autosomal dominant 53 (MRD53)
A disorder characterized by significantly below average general intellectual functioning associated with impairments in adaptive behavior and manifested during the developmental period. {ECO:0000269|PubMed:25533962, ECO:0000269|PubMed:28130356, ECO:0000269|PubMed:29100089, ECO:0000269|PubMed:29560374}. Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 618095]: Intellectual developmental disorder, autosomal recessive 63 (MRT63)
A disorder characterized by significantly below average general intellectual functioning associated with impairments in adaptive behavior and manifested during the developmental period. MRT63 patients manifest global developmental delay, severe intellectual disability, and seizures. {ECO:0000269|PubMed:29784083}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A disorder characterized by significantly below average general intellectual functioning associated with impairments in adaptive behavior and manifested during the developmental period. {ECO:0000269|PubMed:25533962, ECO:0000269|PubMed:28130356, ECO:0000269|PubMed:29100089, ECO:0000269|PubMed:29560374}. Note=The disease is caused by variants affecting the gene represented in this entry.
- A disorder characterized by significantly below average general intellectual functioning associated with impairments in adaptive behavior and manifested during the developmental period. MRT63 patients manifest global developmental delay, severe intellectual disability, and seizures. {ECO:0000269|PubMed:29784083}. Note=The disease is caused by variants affecting the gene represented in this entry.
9 regional properties for Q9UQM7
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Ubiquitin-like domain | 30 - 101 | IPR000626 |
| domain | IBR domain | 334 - 394 | IPR002867-1 |
| domain | IBR domain | 419 - 475 | IPR002867-2 |
| domain | RING/Ubox-like zinc-binding domain | 249 - 339 | IPR041170 |
| domain | Parkin, RING/Ubox like zinc-binding domain | 158 - 236 | IPR041565 |
| domain | TRIAD supradomain | 255 - 482 | IPR044066 |
| domain | E3 ubiquitin-protein ligase parkin, BRcat domain | 322 - 398 | IPR047534 |
| domain | E3 ubiquitin-protein ligase parkin, RING finger, HC subclass | 257 - 315 | IPR047535 |
| domain | E3 ubiquitin-protein ligase parkin, Rcat domain | 428 - 481 | IPR047536 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.17 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24347 | SERINE/THREONINE-PROTEIN KINASE |
| PANTHER Subfamily | PTHR24347:SF384 | CALCIUM_CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II SUBUNIT ALPHA |
| PANTHER Protein Class |
non-receptor serine/threonine protein kinase
protein modifying enzyme |
|
| PANTHER Pathway Category |
Inflammation mediated by chemokine and cytokine signaling pathway CaMK Ionotropic glutamate receptor pathway CaMKII |
|
11 GO annotations of cellular component
| Name | Definition |
|---|---|
| anchoring junction | A cell junction that mechanically attaches a cell (and its cytoskeleton) to neighboring cells or to the extracellular matrix. |
| calcium- and calmodulin-dependent protein kinase complex | An enzyme complex which in eukaryotes is composed of four different chains: alpha, beta, gamma, and delta. The different isoforms assemble into homo- or heteromultimeric holoenzymes composed of 8 to 12 subunits. Catalyzes the phosphorylation of proteins to O-phosphoproteins. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| endocytic vesicle membrane | The lipid bilayer surrounding an endocytic vesicle. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
10 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein kinase activity | Calmodulin-dependent catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; and ATP + a protein threonine = ADP + protein threonine phosphate. |
| glutamate receptor binding | Binding to a glutamate receptor. |
| identical protein binding | Binding to an identical protein or proteins. |
| kinase activity | Catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule. |
| metal ion binding | Binding to a metal ion. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
18 GO annotations of biological process
| Name | Definition |
|---|---|
| angiotensin-activated signaling pathway | A G protein-coupled receptor signaling pathway initiated by angiotensin II binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| calcium ion transport | The directed movement of calcium (Ca) ions into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| dendritic spine development | The process whose specific outcome is the progression of the dendritic spine over time, from its formation to the mature structure. A dendritic spine is a protrusion from a dendrite and a specialized subcellular compartment involved in synaptic transmission. |
| G1/S transition of mitotic cell cycle | The mitotic cell cycle transition by which a cell in G1 commits to S phase. The process begins with the build up of G1 cyclin-dependent kinase (G1 CDK), resulting in the activation of transcription of G1 cyclins. The process ends with the positive feedback of the G1 cyclins on the G1 CDK which commits the cell to S phase, in which DNA replication is initiated. |
| negative regulation of hydrolase activity | Any process that stops or reduces the rate of hydrolase activity, the catalysis of the hydrolysis of various bonds. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| peptidyl-threonine autophosphorylation | The phosphorylation by a protein of one or more of its own threonine amino acid residues, or a threonine residue on an identical protein. |
| positive regulation of calcium ion transport | Any process that activates or increases the frequency, rate or extent of the directed movement of calcium ions into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| positive regulation of cardiac muscle cell apoptotic process | Any process that increases the rate or extent of cardiac cell apoptotic process, a form of programmed cell death induced by external or internal signals that trigger the activity of proteolytic caspases whose actions dismantle a cardiac muscle cell and result in its death. |
| positive regulation of NF-kappaB transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of the transcription factor NF-kappaB. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of endocannabinoid signaling pathway | Any process that modulates the frequency, rate or extent of endocannabinoid signaling pathway. |
| regulation of mitochondrial membrane permeability involved in apoptotic process | Any regulation of mitochondrial membrane permeability that is involved in apoptotic process. |
| regulation of neuron migration | Any process that modulates the frequency, rate or extent of neuron migration. |
| regulation of neuronal synaptic plasticity | A process that modulates neuronal synaptic plasticity, the ability of neuronal synapses to change as circumstances require. They may alter function, such as increasing or decreasing their sensitivity, or they may increase or decrease in actual numbers. |
| regulation of neurotransmitter secretion | Any process that modulates the frequency, rate or extent of the regulated release of a neurotransmitter from a cell. |
| response to ischemia | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a inadequate blood supply. |
49 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3MHJ9 | CAMK2B | Calcium/calmodulin-dependent protein kinase type II subunit beta | Bos taurus (Bovine) | SS |
| Q2HJF7 | CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta | Bos taurus (Bovine) | SS |
| Q5ZKI0 | CAMK2D | Calcium/calmodulin-dependent protein kinase type II delta chain | Gallus gallus (Chicken) | SS |
| Q24210 | CASK | Peripheral plasma membrane protein CASK | Drosophila melanogaster (Fruit fly) | SS |
| Q00168 | CaMKII | Calcium/calmodulin-dependent protein kinase type II alpha chain | Drosophila melanogaster (Fruit fly) | SS |
| O14936 | CASK | Peripheral plasma membrane protein CASK | Homo sapiens (Human) | EV |
| Q16816 | PHKG1 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform | Homo sapiens (Human) | PR |
| P15735 | PHKG2 | Phosphorylase b kinase gamma catalytic chain, liver/testis isoform | Homo sapiens (Human) | PR |
| Q13554 | CAMK2B | Calcium/calmodulin-dependent protein kinase type II subunit beta | Homo sapiens (Human) | EV |
| Q13555 | CAMK2G | Calcium/calmodulin-dependent protein kinase type II subunit gamma | Homo sapiens (Human) | EV |
| Q13557 | CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta | Homo sapiens (Human) | EV |
| Q9H1R3 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Homo sapiens (Human) | EV |
| Q32MK0 | MYLK3 | Myosin light chain kinase 3 | Homo sapiens (Human) | SS |
| Q86YV6 | MYLK4 | Myosin light chain kinase family member 4 | Homo sapiens (Human) | SS |
| P11801 | PSKH1 | Serine/threonine-protein kinase H1 | Homo sapiens (Human) | SS |
| Q6P2M8 | PNCK | Calcium/calmodulin-dependent protein kinase type 1B | Homo sapiens (Human) | SS |
| Q96NX5 | CAMK1G | Calcium/calmodulin-dependent protein kinase type 1G | Homo sapiens (Human) | SS |
| Q8IU85 | CAMK1D | Calcium/calmodulin-dependent protein kinase type 1D | Homo sapiens (Human) | SS |
| Q8NCB2 | CAMKV | CaM kinase-like vesicle-associated protein | Homo sapiens (Human) | SS |
| Q14012 | CAMK1 | Calcium/calmodulin-dependent protein kinase type 1 | Homo sapiens (Human) | EV |
| O70589 | Cask | Peripheral plasma membrane protein CASK | Mus musculus (Mouse) | SS |
| Q923T9 | Camk2g | Calcium/calmodulin-dependent protein kinase type II subunit gamma | Mus musculus (Mouse) | SS |
| Q6PHZ2 | Camk2d | Calcium/calmodulin-dependent protein kinase type II subunit delta | Mus musculus (Mouse) | SS |
| P28652 | Camk2b | Calcium/calmodulin-dependent protein kinase type II subunit beta | Mus musculus (Mouse) | SS |
| P11798 | Camk2a | Calcium/calmodulin-dependent protein kinase type II subunit alpha | Mus musculus (Mouse) | PR |
| Q95266 | CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta | Sus scrofa (Pig) | SS |
| P15791 | Camk2d | Calcium/calmodulin-dependent protein kinase type II subunit delta | Rattus norvegicus (Rat) | SS |
| P11730 | Camk2g | Calcium/calmodulin-dependent protein kinase type II subunit gamma | Rattus norvegicus (Rat) | SS |
| P11275 | Camk2a | Calcium/calmodulin-dependent protein kinase type II subunit alpha | Rattus norvegicus (Rat) | PR |
| P54936 | lin-2 | Protein lin-2 | Caenorhabditis elegans | SS |
| O62305 | unc-43 | Calcium/calmodulin-dependent protein kinase type II | Caenorhabditis elegans | EV |
| Q9ZV15 | CPK20 | Calcium-dependent protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJ61 | CPK25 | Calcium-dependent protein kinase 25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q06850 | CPK1 | Calcium-dependent protein kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38870 | CPK2 | Calcium-dependent protein kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E9C0 | CPK34 | Calcium-dependent protein kinase 34 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FMP5 | CPK17 | Calcium-dependent protein kinase 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42479 | CPK3 | Calcium-dependent protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38872 | CPK6 | Calcium-dependent protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZSA4 | CPK27 | Calcium-dependent protein kinase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39016 | CPK11 | Calcium-dependent protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38869 | CPK4 | Calcium-dependent protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42396 | CPK12 | Calcium-dependent protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q7XJR9 | CPK16 | Calcium-dependent protein kinase 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q1PE17 | CPK18 | Calcium-dependent protein kinase 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FKW4 | CPK28 | Calcium-dependent protein kinase 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q6DGS3 | camk2d2 | Calcium/calmodulin-dependent protein kinase type II delta 2 chain | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q6DEH3 | camk2d1 | Calcium/calmodulin-dependent protein kinase type II delta 1 chain | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MATITCTRFT | EEYQLFEELG | KGAFSVVRRC | VKVLAGQEYA | AKIINTKKLS | ARDHQKLERE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ARICRLLKHP | NIVRLHDSIS | EEGHHYLIFD | LVTGGELFED | IVAREYYSEA | DASHCIQQIL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EAVLHCHQMG | VVHRDLKPEN | LLLASKLKGA | AVKLADFGLA | IEVEGEQQAW | FGFAGTPGYL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SPEVLRKDPY | GKPVDLWACG | VILYILLVGY | PPFWDEDQHR | LYQQIKAGAY | DFPSPEWDTV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TPEAKDLINK | MLTINPSKRI | TAAEALKHPW | ISHRSTVASC | MHRQETVDCL | KKFNARRKLK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GAILTTMLAT | RNFSGGKSGG | NKKSDGVKES | SESTNTTIED | EDTKVRKQEI | IKVTEQLIEA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ISNGDFESYT | KMCDPGMTAF | EPEALGNLVE | GLDFHRFYFE | NLWSRNSKPV | HTTILNPHIH |
| 430 | 440 | 450 | 460 | 470 | |
| LMGDESACIA | YIRITQYLDA | GGIPRTAQSE | ETRVWHRRDG | KWQIVHFHRS | GAPSVLPH |