Q9ULV8
Gene name |
CBLC (CBL3, RNF57) |
Protein name |
E3 ubiquitin-protein ligase CBL-C |
Names |
EC 2.3.2.27 , RING finger protein 57 , RING-type E3 ubiquitin transferase CBL-C , SH3-binding protein CBL-3 , SH3-binding protein CBL-C , Signal transduction protein CBL-C |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:23624 |
EC number |
2.3.2.27: Aminoacyltransferases |
Protein Class |
|
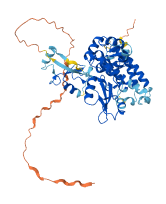
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
9-323 (TKBD) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
11-321 (TKB domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
References
- Dou H et al. (2012) "Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl", Nature structural & molecular biology, 19, 184-92
- Wybenga-Groot LE et al. (2021) "SLAP2 Adaptor Binding Disrupts c-CBL Autoinhibition to Activate Ubiquitin Ligase Function", Journal of molecular biology, 433, 166880
- Kobashigawa Y et al. (2011) "Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b", Proceedings of the National Academy of Sciences of the United States of America, 108, 20579-84
Autoinhibited structure

Activated structure

580 variants for Q9ULV8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1441915381 | 3 | L>V | No |
TOPMed gnomAD |
|
| rs772312798 | 4 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs746098134 | 4 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs772312798 | 4 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs780884827 | 5 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1032029318 | 6 | A>S | No |
TOPMed gnomAD |
|
|
rs1032029318 COSM4079199 |
6 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1837663691 | 6 | A>V | No | TOPMed | |
| rs34932144 | 8 | W>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2098979082 | 8 | W>* | No | gnomAD | |
| rs34932144 | 8 | W>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1265333098 | 11 | Q>* | No |
TOPMed gnomAD |
|
| rs1599853389 | 12 | W>C | No | Ensembl | |
| rs773289730 | 12 | W>G | No |
ExAC TOPMed gnomAD |
|
| rs762946724 | 13 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs762946724 | 13 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs902236013 | 14 | E>D | No |
TOPMed gnomAD |
|
|
rs770372319 COSM4827065 |
14 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1967610568 | 15 | A>V | No | gnomAD | |
| rs1399017208 | 16 | R>C | No |
TOPMed gnomAD |
|
| rs1331119853 | 16 | R>L | No |
TOPMed gnomAD |
|
| rs773891788 | 17 | A>P | No |
ExAC gnomAD |
|
| TCGA novel | 17 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759152258 | 17 | A>V | No |
ExAC gnomAD |
|
| rs1369363757 | 20 | R>Q | No |
TOPMed gnomAD |
|
| rs1967611091 | 20 | R>W | No | gnomAD | |
| rs1000666675 | 23 | R>M | No | TOPMed | |
| rs1225687611 | 24 | M>V | No |
TOPMed gnomAD |
|
| rs767195128 | 25 | L>P | No |
ExAC gnomAD |
|
| rs1341836439 | 26 | Q>* | No |
TOPMed gnomAD |
|
| rs1366818760 | 26 | Q>H | No |
TOPMed gnomAD |
|
| rs200348346 | 26 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs761041888 | 27 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs761041888 | 27 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1418197452 | 27 | R>S | No | gnomAD | |
| rs757709044 | 30 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs778906584 | 31 | Q>* | No |
ExAC gnomAD |
|
| rs778906584 | 31 | Q>E | No |
ExAC gnomAD |
|
| rs778906584 | 31 | Q>K | No |
ExAC gnomAD |
|
| rs750480822 | 31 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs758611319 | 32 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs758611319 | 32 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs866606097 | 34 | D>A | No | gnomAD | |
| rs866606097 | 34 | D>G | No | gnomAD | |
| rs780339606 | 34 | D>N | No |
ExAC gnomAD |
|
| rs1323687243 | 36 | R>Q | No | gnomAD | |
| rs1396002600 | 37 | L>Q | No |
TOPMed gnomAD |
|
| rs1396002600 | 37 | L>R | No |
TOPMed gnomAD |
|
| rs1239073449 | 38 | S>F | No |
TOPMed gnomAD |
|
| rs1967614077 | 38 | S>P | No |
TOPMed gnomAD |
|
| rs749288188 | 39 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs749288188 | 39 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1967614459 | 40 | S>N | No | TOPMed | |
| rs1397115734 | 41 | P>R | No | TOPMed | |
| rs1445083270 | 41 | P>S | No |
TOPMed gnomAD |
|
| rs1445083270 | 41 | P>T | No |
TOPMed gnomAD |
|
| rs983411997 | 42 | P>L | No | TOPMed | |
| rs147885389 | 42 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1967615140 COSM3103348 |
43 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic gnomAD |
| rs1967615300 | 44 | L>Q | No | TOPMed | |
|
rs931299718 COSM3971083 |
45 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1967615618 | 46 | D>N | No |
TOPMed gnomAD |
|
| rs775127808 | 49 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs148948759 | 49 | P>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs148948759 | 49 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs775127808 | 49 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1446927956 | 50 | R>G | No | gnomAD | |
| rs776955522 | 50 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs762265193 | 51 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1163724186 | 53 | Q>K | No | gnomAD | |
| COSM3535746 | 56 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765766542 | 56 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs765766542 | 56 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1967616911 | 57 | E>A | No |
TOPMed gnomAD |
|
| rs1455126115 | 57 | E>D | No | gnomAD | |
| rs943884863 | 57 | E>Q | No | TOPMed | |
| rs572115097 | 58 | V>A | No |
1000Genomes ExAC gnomAD |
|
| rs541721336 | 59 | A>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1389009213 | 59 | A>T | No | gnomAD | |
| rs541721336 | 59 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1320747390 | 61 | S>P | No | gnomAD | |
| rs1967617790 | 62 | R>P | No | TOPMed | |
| TCGA novel | 62 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1967617699 | 62 | R>W | No | Ensembl | |
| rs777753616 | 63 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs777753616 | 63 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs115775900 | 63 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM4079200 | 64 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1967618769 | 66 | G>D | No | Ensembl | |
| rs1226709242 | 66 | G>R | No |
TOPMed gnomAD |
|
| rs1226709242 | 66 | G>S | No |
TOPMed gnomAD |
|
| rs1159769841 | 67 | G>* | No | Ensembl | |
| rs749090451 | 67 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1330131412 | 68 | G>A | No |
TOPMed gnomAD |
|
| rs1967619043 | 68 | G>R | No | Ensembl | |
| rs1207100126 | 69 | G>S | No | gnomAD | |
| rs778923243 | 71 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs757201148 | 71 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs778923243 | 71 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1967619915 | 72 | G>C | No | gnomAD | |
| rs949078334 | 72 | G>D | No | gnomAD | |
| COSM3892838 | 72 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1180878122 | 74 | G>R | No | gnomAD | |
| rs1180878122 | 74 | G>S | No | gnomAD | |
| rs1436824407 | 75 | G>C | No | TOPMed | |
| rs1436824407 | 75 | G>R | No | TOPMed | |
| rs1967620589 | 76 | S>F | No | TOPMed | |
| rs1967620512 | 76 | S>P | No | TOPMed | |
| rs530433110 | 78 | D>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1426068218 | 78 | D>N | No |
TOPMed gnomAD |
|
| rs530433110 | 78 | D>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1455042710 | 79 | F>V | No | gnomAD | |
| rs1000593338 | 81 | L>F | No |
TOPMed gnomAD |
|
| rs1000593338 | 81 | L>V | No |
TOPMed gnomAD |
|
| rs1032038079 | 83 | Y>C | No |
TOPMed gnomAD |
|
| rs1967621273 | 83 | Y>D | No | TOPMed | |
| rs1032038079 | 83 | Y>S | No |
TOPMed gnomAD |
|
| rs1260448016 | 85 | A>D | No |
TOPMed gnomAD |
|
| rs1967621647 | 86 | N>K | No | Ensembl | |
| rs1212319676 | 86 | N>S | No |
TOPMed gnomAD |
|
| rs746709240 | 87 | L>V | No |
ExAC gnomAD |
|
| rs1967621877 | 88 | E>G | No | Ensembl | |
| rs1377862471 | 89 | A>D | No |
TOPMed gnomAD |
|
| rs1277462108 | 89 | A>T | No |
TOPMed gnomAD |
|
| rs548486447 | 90 | K>E | No | gnomAD | |
| rs768263588 | 90 | K>R | No |
ExAC gnomAD |
|
| rs1336377385 | 91 | S>N | No | gnomAD | |
| rs1344052233 | 93 | Q>H | No |
TOPMed gnomAD |
|
| rs1967622995 | 95 | A>G | No | gnomAD | |
| rs1599853907 | 96 | A>T | No | TOPMed | |
| rs1967623248 | 99 | P>A | No | Ensembl | |
|
COSM3535748 rs1568554125 |
100 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs967594513 | 101 | R>G | No |
TOPMed gnomAD |
|
| rs1401164342 | 101 | R>P | No |
TOPMed gnomAD |
|
| rs1401164342 | 101 | R>Q | No |
TOPMed gnomAD |
|
| rs967594513 | 101 | R>W | No |
TOPMed gnomAD |
|
| rs765456312 | 102 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1204290245 | 103 | R>* | No | gnomAD | |
|
COSM3535749 rs1035887672 |
104 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs1251944026 | 104 | R>S | No | gnomAD | |
| rs1035887672 | 104 | R>T | No | TOPMed | |
| rs1030666838 | 105 | S>G | No | Ensembl | |
| rs1568554186 | 106 | A>T | No | gnomAD | |
| rs1967624562 | 107 | N>D | No | TOPMed | |
| rs563733027 | 108 | D>E | No |
1000Genomes TOPMed gnomAD |
|
| rs1967624648 | 108 | D>N | No | Ensembl | |
| rs773804002 | 109 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1196319108 | 109 | E>K | No |
TOPMed gnomAD |
|
| rs1196319108 | 109 | E>Q | No |
TOPMed gnomAD |
|
| rs1967624966 | 109 | E>V | No | Ensembl | |
| rs1421419024 | 110 | L>F | No |
TOPMed gnomAD |
|
| rs919974224 | 110 | L>P | No |
TOPMed gnomAD |
|
| COSM3990103 | 111 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs983465843 | 111 | F>S | No |
TOPMed gnomAD |
|
| rs1174706035 | 112 | R>P | No | gnomAD | |
| rs1377633149 | 113 | A>E | No |
TOPMed gnomAD |
|
| rs1967625763 | 114 | G>S | No | Ensembl | |
| rs952747717 | 115 | S>F | No |
TOPMed gnomAD |
|
| rs1014912119 | 115 | S>P | No |
TOPMed gnomAD |
|
| rs1014912119 | 115 | S>T | No |
TOPMed gnomAD |
|
| rs1967626328 | 117 | L>R | No | TOPMed | |
| rs1465638279 | 117 | L>V | No |
TOPMed gnomAD |
|
| rs1967626391 | 118 | R>T | No | TOPMed | |
| rs372975672 | 119 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs201563104 COSM998051 |
119 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs935339144 | 120 | Q>* | No | TOPMed | |
| rs1967706738 | 120 | Q>H | No | gnomAD | |
| rs2965127 | 121 | L>V | No | Ensembl | |
| rs142893738 | 122 | A>S | No |
ESP TOPMed gnomAD |
|
| rs2122390654 | 123 | K>N | No | Ensembl | |
| rs774630684 | 123 | K>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 123 | K>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3535750 | 126 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1170657904 | 129 | S>N | No | gnomAD | |
| rs1599857318 | 130 | H>R | No | Ensembl | |
| rs1967707940 | 131 | M>I | No | TOPMed | |
| rs140833100 | 132 | H>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs199953740 | 133 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199953740 | 133 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs765128314 | 134 | E>K | No |
ExAC gnomAD |
|
| rs1967708704 | 136 | H>Y | No | TOPMed | |
| rs1289276617 | 137 | A>P | No |
TOPMed gnomAD |
|
| rs1289276617 | 137 | A>T | No |
TOPMed gnomAD |
|
| rs376312238 | 137 | A>V | No |
TOPMed gnomAD |
|
| rs1394255813 | 138 | L>F | No |
TOPMed gnomAD |
|
| rs1394255813 | 138 | L>V | No |
TOPMed gnomAD |
|
| rs981330867 | 139 | F>L | No | gnomAD | |
| rs865916210 | 140 | P>A | No | TOPMed | |
| rs751438592 | 140 | P>L | No |
ExAC gnomAD |
|
| rs865916210 | 140 | P>S | No | TOPMed | |
| rs1967709870 | 141 | G>E | No | TOPMed | |
| rs377440313 | 141 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 141 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747887583 | 142 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs747887583 | 142 | G>V | No |
ExAC gnomAD |
|
| rs144857168 | 143 | K>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs144857168 | 143 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1217917009 | 143 | K>M | No | gnomAD | |
| rs1967710418 | 144 | Y>N | No | gnomAD | |
| rs1261713017 | 144 | Y>S | No | gnomAD | |
| rs1487014571 | 146 | G>E | No | gnomAD | |
| rs771366662 | 147 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs749602901 | 147 | H>Y | No |
ExAC gnomAD |
|
| rs1409817102 | 148 | M>I | No |
TOPMed gnomAD |
|
|
COSM4079201 rs1190196751 |
148 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1967711103 | 148 | M>T | No | Ensembl | |
| rs775004600 | 150 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1227040829 | 150 | Q>R | No |
TOPMed gnomAD |
|
| rs1346752627 | 152 | T>I | No | gnomAD | |
| rs1568555756 | 152 | T>P | No | Ensembl | |
| rs1967711832 | 153 | K>E | No | Ensembl | |
| rs35106910 | 153 | K>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs35106910 | 153 | K>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs775409459 | 154 | A>P | No |
ExAC gnomAD |
|
| rs1283928118 | 155 | P>T | No | TOPMed | |
| rs764214326 | 156 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs371124704 | 158 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs371124704 | 158 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs762748099 | 159 | F>C | No |
ExAC gnomAD |
|
| rs1795223132 | 164 | C>F | No |
TOPMed gnomAD |
|
| rs1967713309 | 164 | C>G | No | TOPMed | |
| rs1384229661 | 165 | G>E | No |
TOPMed gnomAD |
|
| rs140442555 | 165 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs752145860 | 167 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs201807125 | 167 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs752145860 | 167 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs746229759 | 168 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs1423933513 | 169 | V>A | No | gnomAD | |
| rs1967720004 | 169 | V>M | No |
TOPMed gnomAD |
|
| rs1295857633 | 170 | L>M | No | gnomAD | |
| rs1967720388 | 171 | P>S | No | TOPMed | |
| rs2122394590 | 174 | E>* | No | Ensembl | |
| rs114569424 | 178 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1383499306 | 178 | L>H | No |
TOPMed gnomAD |
|
| rs1967720892 | 180 | G>S | No | TOPMed | |
| rs137908794 | 181 | T>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1599857841 | 181 | T>P | No | Ensembl | |
| rs200445340 | 185 | V>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200445340 | 185 | V>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2122394810 | 185 | V>M | No | Ensembl | |
| COSM3535751 | 186 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1274103082 | 187 | P>R | No | gnomAD | |
| rs770003480 | 188 | G>A | No |
ExAC gnomAD |
|
| rs1399203802 | 188 | G>R | No | gnomAD | |
| rs759339597 | 190 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs944222784 | 190 | T>P | No | Ensembl | |
| rs759339597 | 190 | T>R | No |
ExAC TOPMed gnomAD |
|
| COSM3535752 | 191 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1188155125 | 192 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1328874977 | 192 | L>V | No | TOPMed | |
| rs775321656 | 194 | L>* | No |
ExAC gnomAD |
|
| rs775321656 | 194 | L>S | No |
ExAC gnomAD |
|
| rs372310813 | 195 | R>C | No |
ESP ExAC gnomAD |
|
| rs372310813 | 195 | R>G | No |
ESP ExAC gnomAD |
|
| rs575600792 | 195 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs149481548 | 196 | T>I | No |
ESP TOPMed gnomAD |
|
| rs2122395288 | 197 | T>A | No | Ensembl | |
| rs868021585 | 198 | I>T | No |
TOPMed gnomAD |
|
| rs764965728 | 199 | D>Y | No |
ExAC gnomAD |
|
| rs1326165886 | 201 | T>P | No | gnomAD | |
| rs750033744 | 201 | T>S | No |
ExAC gnomAD |
|
| rs1272373909 | 202 | C>Y | No | gnomAD | |
| rs1232819921 | 203 | S>G | No |
TOPMed gnomAD |
|
| rs780423177 | 204 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs755482665 | 205 | H>Y | No |
ExAC gnomAD |
|
| rs781320503 | 206 | V>M | No |
ExAC gnomAD |
|
| rs769807888 | 210 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs769807888 | 210 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs749488002 | 212 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs749488002 | 212 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs760567497 | 213 | V>D | No |
ExAC gnomAD |
|
|
rs557856786 COSM4382626 |
213 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| COSM474903 | 213 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768709346 | 214 | F>L | No | ExAC | |
| rs1967725365 | 215 | T>P | No | TOPMed | |
| rs776338309 | 217 | L>F | No |
ExAC gnomAD |
|
| rs761418172 | 217 | L>P | No | ExAC | |
| rs1967725891 | 218 | F>V | No | Ensembl | |
| TCGA novel | 220 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759899304 | 221 | W>* | No |
ExAC gnomAD |
|
| rs1967772777 | 223 | T>A | No | Ensembl | |
| rs1483170598 | 223 | T>I | No | TOPMed | |
| rs953975785 | 224 | L>H | No | Ensembl | |
| rs1418805363 | 227 | N>S | No |
TOPMed gnomAD |
|
| rs767829298 | 228 | W>S | No |
ExAC gnomAD |
|
| rs753226393 | 230 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs753226393 | 230 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs373513195 | 232 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1204850508 | 234 | N>H | No | TOPMed | |
|
COSM1394498 COSM5135618 |
236 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764196556 | 236 | P>T | No |
ExAC gnomAD |
|
| rs753946761 | 237 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1324057395 | 238 | Y>C | No | gnomAD | |
| rs757478241 | 238 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs2122409934 | 239 | M>K | No | Ensembl | |
| rs1369996784 | 239 | M>V | No | gnomAD | |
| rs1321205852 | 240 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1291457488 | 242 | L>R | No | TOPMed | |
| rs779208700 | 245 | D>G | No |
ExAC gnomAD |
|
| rs779208700 | 245 | D>V | No |
ExAC gnomAD |
|
| rs1599859761 | 246 | E>K | No | Ensembl | |
|
COSM5833772 COSM3823385 |
247 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs148127553 COSM3103362 |
247 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| COSM998053 | 247 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1226540961 | 248 | Q>P | No | gnomAD | |
| rs781082374 | 249 | E>G | No | ExAC | |
| rs141956736 | 250 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs147316311 | 250 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs147316311 | 250 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs141956736 | 250 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1392563549 | 252 | Q>P | No | Ensembl | |
| rs1392563549 | 252 | Q>R | No | Ensembl | |
| rs138510701 | 254 | C>F | No |
1000Genomes ExAC gnomAD |
|
| rs1269891319 | 255 | R>G | No | gnomAD | |
| rs759141229 | 256 | D>E | No |
ExAC gnomAD |
|
| TCGA novel | 256 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 257 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1967776739 | 257 | K>E | No | Ensembl | |
| rs1463540514 | 259 | G>D | No | gnomAD | |
| rs767920193 | 259 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1162871941 | 260 | S>T | No | gnomAD | |
| rs1967826418 | 261 | Y>F | No | TOPMed | |
| rs1489569438 | 263 | F>L | No | gnomAD | |
| rs761040194 | 264 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1568558009 | 264 | R>W | No | TOPMed | |
| rs777222729 | 265 | P>R | No |
ExAC gnomAD |
|
| rs1486005385 | 265 | P>S | No |
TOPMed gnomAD |
|
| rs765404486 | 266 | S>N | No |
ExAC gnomAD |
|
| rs1224709577 | 266 | S>R | No |
TOPMed gnomAD |
|
| rs558587668 | 266 | S>R | No |
1000Genomes ExAC gnomAD |
|
| rs1350362593 | 267 | C>R | No |
TOPMed gnomAD |
|
| rs1350362593 | 267 | C>S | No |
TOPMed gnomAD |
|
|
rs750497513 COSM998055 |
269 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs201207715 | 269 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201207715 | 269 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs780929631 | 270 | L>Q | No | Ensembl | |
| rs995111410 | 271 | G>E | No |
TOPMed gnomAD |
|
| rs751768100 | 271 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1295944061 | 272 | Q>* | No | gnomAD | |
| rs1967828297 | 272 | Q>R | No | TOPMed | |
| rs1351479922 | 273 | W>* | No | Ensembl | |
| rs1967828447 | 274 | A>P | No | Ensembl | |
| rs753864374 | 275 | I>M | No |
ExAC gnomAD |
|
| rs777383264 | 275 | I>N | No |
ExAC gnomAD |
|
| rs755972476 | 275 | I>V | No |
ExAC gnomAD |
|
| rs778549620 | 276 | G>A | No |
ExAC gnomAD |
|
| rs143342303 | 276 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1280307994 | 277 | Y>C | No |
TOPMed gnomAD |
|
| rs1967829312 | 278 | V>A | No | Ensembl | |
| rs1399897724 | 278 | V>L | No | TOPMed | |
| rs1405445288 | 279 | S>N | No |
TOPMed gnomAD |
|
| rs745367240 | 280 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs1967829584 | 281 | D>H | No | Ensembl | |
| rs1231229315 | 283 | S>N | No | gnomAD | |
| rs1239786858 | 287 | T>A | No | gnomAD | |
| rs1251878645 | 287 | T>I | No |
TOPMed gnomAD |
|
| rs1452277500 | 288 | I>T | No | gnomAD | |
| rs1967830276 | 289 | P>S | No | Ensembl | |
| rs746620893 | 290 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1395689292 | 291 | N>K | No | gnomAD | |
| rs769084079 | 293 | P>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 293 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1416071534 | 293 | P>T | No | gnomAD | |
| rs762419781 | 297 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs1438693175 | 298 | L>V | No | gnomAD | |
| rs1967831293 | 301 | G>V | No | Ensembl | |
| rs1439241795 | 302 | Q>* | No | gnomAD | |
| rs370388709 | 304 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs773800624 | 304 | D>Y | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 305 | G>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs61755281 | 305 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs147455821 | 308 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs147455821 | 308 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs780714860 | 311 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2122472012 | 313 | K>E | No | Ensembl | |
| rs1284066266 | 313 | K>R | No | gnomAD | |
| rs1968005839 | 314 | T>P | No | Ensembl | |
| rs747797534 | 315 | H>Y | No |
ExAC gnomAD |
|
| rs756569446 | 316 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs182254561 | 318 | D>Y | No | 1000Genomes | |
| rs1258130935 | 322 | L>F | No | gnomAD | |
| rs760486029 | 323 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs745945780 | 325 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs745945780 | 325 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs774632827 | 325 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs745945780 | 325 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs772120427 | 326 | E>D | No |
ExAC gnomAD |
|
| rs1968007330 | 327 | P>L | No | TOPMed | |
| rs2122472442 | 327 | P>S | No | Ensembl | |
| rs760898443 | 330 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs139273613 | 330 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs139273613 | 330 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs772959528 | 331 | I>N | No |
ExAC gnomAD |
|
| rs766142122 | 332 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs536360053 | 332 | H>R | No |
1000Genomes ExAC gnomAD |
|
| rs1968008177 | 333 | V>A | No |
TOPMed gnomAD |
|
| rs751449455 | 333 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1968008270 | 335 | E>K | No | TOPMed | |
| rs1166415820 | 336 | E>D | No |
TOPMed gnomAD |
|
| rs1279432747 | 336 | E>G | No | gnomAD | |
| rs897758747 | 336 | E>K | No | gnomAD | |
| rs758907585 | 338 | L>V | No | ExAC | |
| rs1458790653 | 339 | Q>* | No | gnomAD | |
| rs1968075858 | 339 | Q>R | No | TOPMed | |
| rs1316693211 | 340 | L>F | No |
TOPMed gnomAD |
|
| rs961495774 | 341 | Y>C | No |
TOPMed gnomAD |
|
| rs961495774 | 341 | Y>F | No |
TOPMed gnomAD |
|
| rs2122488341 | 342 | W>C | No | Ensembl | |
| rs747076535 | 343 | A>V | No |
ExAC gnomAD |
|
| rs149074838 | 344 | M>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2122488403 | 345 | D>N | No | Ensembl | |
| rs1240356013 | 346 | S>F | No | TOPMed | |
| rs748321868 | 347 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs548133945 | 347 | T>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs748321868 | 347 | T>P | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 349 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1236477787 | 350 | L>F | No | TOPMed | |
| rs759463367 | 350 | L>H | No |
ExAC gnomAD |
|
| rs759463367 | 350 | L>P | No |
ExAC gnomAD |
|
| rs1236477787 | 350 | L>V | No | TOPMed | |
| rs775236973 | 351 | C>R | No |
ExAC gnomAD |
|
| rs760268560 | 352 | K>E | No |
ExAC TOPMed gnomAD |
|
|
rs2122488661 TCGA novel |
352 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1303390345 | 354 | C>* | No | gnomAD | |
| rs142187731 | 354 | C>S | No |
1000Genomes TOPMed |
|
| rs142187731 | 354 | C>Y | No |
1000Genomes TOPMed |
|
| rs952137350 | 355 | A>D | No |
TOPMed gnomAD |
|
| rs952137350 | 355 | A>G | No |
TOPMed gnomAD |
|
| rs952137350 | 355 | A>V | No |
TOPMed gnomAD |
|
| rs1968078232 | 356 | E>A | No | Ensembl | |
|
COSM4834391 rs763654909 |
356 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs763654909 | 356 | E>Q | No |
ExAC gnomAD |
|
| rs761530274 | 357 | S>C | No |
ExAC gnomAD |
|
| rs148655912 | 357 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1023454829 | 357 | S>R | No |
TOPMed gnomAD |
|
| rs750794901 | 358 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1445991874 | 358 | N>S | No | TOPMed | |
| rs1968078892 | 359 | K>M | No | TOPMed | |
| rs758885229 | 360 | D>N | No |
ExAC gnomAD |
|
| rs1968079066 | 360 | D>V | No | gnomAD | |
| COSM3535756 | 361 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs766921120 | 363 | I>N | No |
ExAC gnomAD |
|
| rs766921120 | 363 | I>T | No |
ExAC gnomAD |
|
| rs971614881 | 365 | P>L | No |
TOPMed gnomAD |
|
| rs923737137 | 365 | P>S | No |
TOPMed gnomAD |
|
| rs923737137 | 365 | P>T | No |
TOPMed gnomAD |
|
| rs374379148 | 366 | C>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs374379148 | 366 | C>Y | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4989668 rs377703099 |
367 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1295034727 | 368 | H>Y | No | gnomAD | |
| rs1381529020 | 370 | L>F | No | gnomAD | |
| TCGA novel | 370 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1968080800 | 375 | L>P | No | TOPMed | |
| rs1968080800 | 375 | L>Q | No | TOPMed | |
| rs1321944719 | 376 | A>T | No | gnomAD | |
| rs768327321 | 377 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs768327321 | 377 | A>S | No |
ExAC TOPMed gnomAD |
|
| COSM4829164 | 378 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1349195973 | 379 | Q>* | No | gnomAD | |
| rs1348916531 | 380 | H>R | No |
TOPMed gnomAD |
|
| rs753138447 | 381 | S>L | No |
ExAC gnomAD |
|
| rs1302998194 | 381 | S>P | No |
TOPMed gnomAD |
|
| COSM4079203 | 385 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1310418136 | 385 | T>N | No |
TOPMed gnomAD |
|
| rs1232691082 | 387 | P>L | No |
TOPMed gnomAD |
|
| rs146735092 | 387 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs757386289 | 388 | F>L | No | ExAC | |
| rs1599871855 | 388 | F>S | No | Ensembl | |
| rs374326666 | 389 | C>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs751385301 | 390 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs754761911 COSM3103385 |
390 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs751385301 | 390 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1968106082 | 391 | C>F | No | TOPMed | |
| rs35457630 | 392 | E>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1968106402 | 392 | E>G | No | TOPMed | |
| rs35457630 | 392 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1053116620 | 394 | K>E | No | TOPMed | |
| rs769396781 | 394 | K>N | No |
ExAC gnomAD |
|
| rs1968106658 | 396 | W>* | No | Ensembl | |
| rs1239721663 | 396 | W>* | No | gnomAD | |
| TCGA novel | 396 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1192911372 | 397 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 397 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777117772 | 398 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs777117772 | 398 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs749036591 | 399 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1968107345 | 400 | S>N | No | Ensembl | |
| COSM69047 | 400 | S>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1425280513 | 401 | I>V | No | gnomAD | |
| rs1167660429 | 402 | Y>* | No |
TOPMed gnomAD |
|
| rs1968107653 | 402 | Y>C | No | TOPMed | |
| rs1434461356 | 402 | Y>H | No | gnomAD | |
| rs1399153594 | 403 | Q>P | No | gnomAD | |
| rs3208856 | 405 | H>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2122494882 | 405 | H>P | No | Ensembl | |
|
rs3208856 VAR_018298 |
405 | H>Y | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs368494441 | 406 | G>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs2054488140 | 406 | G>D | No | Ensembl | |
| rs368494441 | 406 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs368494441 | 406 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1968108316 | 407 | Q>E | No | TOPMed | |
| rs772388756 | 408 | A>P | No |
ExAC gnomAD |
|
| rs772388756 | 408 | A>S | No |
ExAC gnomAD |
|
| rs1568563845 | 413 | S>* | No | TOPMed | |
| rs1568563845 | 413 | S>L | No | TOPMed | |
| rs17852620 | 413 | S>P | No | Ensembl | |
| rs1968108896 | 414 | G>R | No | Ensembl | |
| rs1293493860 | 416 | S>R | No |
TOPMed gnomAD |
|
| rs764271266 | 416 | S>T | No |
ExAC gnomAD |
|
| rs762183942 | 418 | D>N | No |
ExAC gnomAD |
|
| rs765569181 | 419 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs750604085 | 419 | Q>H | No |
ExAC gnomAD |
|
| rs1289790618 | 419 | Q>R | No | TOPMed | |
| rs754849774 | 420 | E>V | No |
ExAC gnomAD |
|
| rs781113299 | 421 | G>S | No |
ExAC gnomAD |
|
| rs994347051 | 423 | E>K | No |
TOPMed gnomAD |
|
| rs1331839617 | 425 | E>K | No | Ensembl | |
| rs1172300265 | 425 | E>V | No |
TOPMed gnomAD |
|
| rs1025942028 | 426 | L>V | No |
TOPMed gnomAD |
|
| rs1404017582 | 427 | G>R | No | gnomAD | |
| rs1187731803 | 429 | V>L | No |
TOPMed gnomAD |
|
| rs1187731803 | 429 | V>M | No |
TOPMed gnomAD |
|
| COSM3892840 | 430 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1474012971 | 430 | P>R | No |
TOPMed gnomAD |
|
| rs745556828 | 432 | S>* | No |
ExAC TOPMed gnomAD |
|
| rs1838178777 | 432 | S>A | No | Ensembl | |
|
COSM5141243 COSM1394500 rs745556828 |
432 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs557733635 | 433 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM3535757 | 434 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs116023028 | 435 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs777033769 | 436 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1265994312 | 437 | P>R | No | gnomAD | |
| rs1599872911 | 438 | P>S | No | Ensembl | |
| rs770035168 | 439 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs374048625 | 439 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1242181381 | 441 | D>Y | No | gnomAD | |
| rs1272578610 | 442 | L>P | No |
TOPMed gnomAD |
|
| rs1447035976 | 443 | P>S | No | TOPMed | |
| rs1280385353 | 444 | P>A | No | gnomAD | |
| rs773422704 | 444 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs773422704 | 444 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1968132604 | 445 | R>K | No | Ensembl | |
| rs766555920 | 445 | R>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1183446053 | 446 | K>N | No | TOPMed | |
| rs1386405463 | 446 | K>R | No |
TOPMed gnomAD |
|
| rs763325522 | 447 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs138476890 | 450 | A>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs138476890 | 450 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs997076042 | 451 | Q>R | No | TOPMed | |
| rs1968133016 | 452 | P>S | No | TOPMed | |
| rs1599872994 | 453 | K>E | No | Ensembl | |
| rs61750956 | 455 | R>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1968269207 | 455 | R>S | No | gnomAD | |
| rs61750956 | 455 | R>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs762400110 | 456 | L>P | No |
ExAC gnomAD |
|
| rs1050232407 | 458 | K>R | No |
TOPMed gnomAD |
|
| rs1470815090 | 461 | S>P | No | TOPMed | |
| rs753128512 | 462 | P>L | No |
ExAC gnomAD |
|
| COSM3535759 | 462 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1968269757 | 462 | P>T | No | Ensembl | |
|
COSM5211983 COSM439764 |
463 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM6085374 COSM6085373 |
464 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756481545 | 464 | A>P | No |
ExAC gnomAD |
|
| rs756481545 | 464 | A>T | No |
ExAC gnomAD |
|
| rs904426529 | 465 | A>V | No | Ensembl | |
| COSM998060 | 467 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1185174270 | 467 | G>E | No | gnomAD | |
| rs1968270219 | 469 | Q>* | No | Ensembl | |
| rs1968270267 | 469 | Q>P | No | gnomAD | |
| rs1968270267 | 469 | Q>R | No | gnomAD | |
| TCGA novel | 470 | D>missing | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1269858060 | 470 | D>G | No | TOPMed | |
| rs770969313 | 470 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs746139762 | 472 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 473 | P>missing | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1418606115 | 473 | P>A | No |
TOPMed gnomAD |
|
| rs1298249941 | 473 | P>L | No | gnomAD | |
| rs1298249941 | 473 | P>R | No | gnomAD | |
| rs1288290895 | 475 | A>C | No | TOPMed | |
| COSM998062 | 475 | A>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
No associated diseases with Q9ULV8
10 regional properties for Q9ULV8
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Death domain | 838 - 929 | IPR000488 |
| repeat | Thrombospondin type-1 (TSP1) repeat | 260 - 314 | IPR000884-1 |
| repeat | Thrombospondin type-1 (TSP1) repeat | 316 - 368 | IPR000884-2 |
| domain | ZU5 domain | 528 - 673 | IPR000906 |
| domain | Immunoglobulin subtype 2 | 179 - 246 | IPR003598 |
| domain | Immunoglobulin subtype | 173 - 258 | IPR003599 |
| domain | Immunoglobulin-like domain | 175 - 256 | IPR007110 |
| domain | Immunoglobulin I-set | 168 - 257 | IPR013098 |
| domain | UPA domain | 678 - 817 | IPR033772 |
| domain | UNC5C, death domain | 844 - 926 | IPR042154 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.27 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| epidermal growth factor receptor binding | Binding to an epidermal growth factor receptor. |
| phosphotyrosine residue binding | Binding to a phosphorylated tyrosine residue within a protein. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| SH3 domain binding | Binding to a SH3 domain (Src homology 3) of a protein, small protein modules containing approximately 50 amino acid residues found in a great variety of intracellular or membrane-associated proteins. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond |
| zinc ion binding | Binding to a zinc ion (Zn). |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| cell surface receptor signaling pathway | The series of molecular signals initiated by activation of a receptor on the surface of a cell. The pathway begins with binding of an extracellular ligand to a cell surface receptor, or for receptors that signal in the absence of a ligand, by ligand-withdrawal or the activity of a constitutively active receptor. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| negative regulation of epidermal growth factor receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of epidermal growth factor receptor signaling pathway activity. |
| negative regulation of epidermal growth factor-activated receptor activity | Any process that stops, prevents, or reduces the frequency, rate or extent of EGF-activated receptor activity. |
| negative regulation of MAP kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of MAP kinase activity. |
| negative regulation of neuron apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process in neurons. |
| protein stabilization | Any process involved in maintaining the structure and integrity of a protein and preventing it from degradation or aggregation. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| response to glial cell derived neurotrophic factor | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a glial cell derived neurotrophic factor stimulus. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of a ubiquitin group, or multiple ubiquitin groups, to the protein. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q13191 | CBLB | E3 ubiquitin-protein ligase CBL-B | Homo sapiens (Human) | EV |
| P22681 | CBL | E3 ubiquitin-protein ligase CBL | Homo sapiens (Human) | EV |
| P22682 | Cbl | E3 ubiquitin-protein ligase CBL | Mus musculus (Mouse) | SS |
| Q3TTA7 | Cblb | E3 ubiquitin-protein ligase CBL-B | Mus musculus (Mouse) | SS |
| Q80XL1 | Cblc | E3 ubiquitin-protein ligase CBL-C | Mus musculus (Mouse) | SS |
| Q8K4S7 | Cblb | E3 ubiquitin-protein ligase CBL-B | Rattus norvegicus (Rat) | SS |
| G3V8H4 | Cblc | E3 ubiquitin-protein ligase CBL-C | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MALAVAPWGR | QWEEARALGR | AVRMLQRLEE | QCVDPRLSVS | PPSLRDLLPR | TAQLLREVAH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SRRAAGGGGP | GGPGGSGDFL | LIYLANLEAK | SRQVAALLPP | RGRRSANDEL | FRAGSRLRRQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LAKLAIIFSH | MHAELHALFP | GGKYCGHMYQ | LTKAPAHTFW | RESCGARCVL | PWAEFESLLG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TCHPVEPGCT | ALALRTTIDL | TCSGHVSIFE | FDVFTRLFQP | WPTLLKNWQL | LAVNHPGYMA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FLTYDEVQER | LQACRDKPGS | YIFRPSCTRL | GQWAIGYVSS | DGSILQTIPA | NKPLSQVLLE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GQKDGFYLYP | DGKTHNPDLT | ELGQAEPQQR | IHVSEEQLQL | YWAMDSTFEL | CKICAESNKD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VKIEPCGHLL | CSCCLAAWQH | SDSQTCPFCR | CEIKGWEAVS | IYQFHGQATA | EDSGNSSDQE |
| 430 | 440 | 450 | 460 | 470 | |
| GRELELGQVP | LSAPPLPPRP | DLPPRKPRNA | QPKVRLLKGN | SPPAALGPQD | PAPA |