Q9ULL4
Gene name |
PLXNB3 (KIAA1206, PLXN6) |
Protein name |
Plexin-B3 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5365 |
EC number |
|
Protein Class |
|
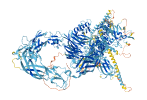
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1327-1876 (Plexin, cytoplasmic RasGAP domain) |
Relief mechanism |
Partner binding |
Assay |
|
Target domain |
1327-1876 (Plexin, cytoplasmic RasGAP domain) |
Relief mechanism |
Partner binding |
Assay |
|
Target domain |
1327-1876 (Plexin, RasGAP domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
- Wang Y et al. (2012) "Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization", Science signaling, 5, ra6
- He H et al. (2009) "Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration", Proceedings of the National Academy of Sciences of the United States of America, 106, 15610-5
- Takahashi T et al. (2001) "Plexina1 autoinhibition by the plexin sema domain", Neuron, 29, 429-39
Autoinhibited structure

Activated structure

1 structures for Q9ULL4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9ULL4-F1 | Predicted | AlphaFoldDB |
1783 variants for Q9ULL4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV002266657 rs2148413029 |
74 | L>F | PLXNB3-related Intellectual disability [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs2091974044 RCV001262161 |
1425 | T>N | Neurodevelopmental disorder [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs148960424 RCV000900405 |
1 | M>V | No |
ClinVar dbSNP |
|
| rs781805567 | 2 | C>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1168379326 | 3 | H>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs782172672 | 4 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2091847954 | 4 | A>V | No |
TOPMed gnomAD |
|
| rs1557058874 | 5 | A>G | No | Ensembl | |
| rs148108922 | 5 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091848053 | 6 | Q>* | No | TOPMed | |
| rs1175080153 | 7 | E>K | No |
1000Genomes TOPMed gnomAD |
|
| rs2091848099 | 9 | P>L | No | Ensembl | |
| rs1432873659 | 10 | L>V | No |
TOPMed gnomAD |
|
| rs1251342410 | 12 | H>Y | No |
TOPMed gnomAD |
|
| rs141960270 | 13 | H>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1458228815 | 15 | M>T | No |
TOPMed gnomAD |
|
| rs1557059395 | 16 | A>D | No | gnomAD | |
| rs782125512 | 16 | A>T | No | ExAC | |
| rs1252299131 | 17 | P>S | No |
TOPMed gnomAD |
|
| rs368049202 | 18 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091864609 | 19 | M>T | No | TOPMed | |
| rs782457646 | 20 | A>P | No |
ExAC gnomAD |
|
| rs782457646 | 20 | A>S | No |
ExAC gnomAD |
|
| rs782457646 | 20 | A>T | No |
ExAC gnomAD |
|
| rs782666089 | 21 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs782666089 | 21 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs782221652 | 21 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1557059415 | 22 | W>C | No |
TOPMed gnomAD |
|
| rs782365591 | 26 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782365591 | 26 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs35537879 RCV000967615 |
28 | C>Y | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs372269937 | 31 | L>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs372269937 | 31 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1409696454 | 36 | P>L | No |
TOPMed gnomAD |
|
| rs1409696454 | 36 | P>R | No |
TOPMed gnomAD |
|
| rs1557059431 | 36 | P>S | No | gnomAD | |
| rs1175052764 | 37 | P>L | No |
TOPMed gnomAD |
|
|
COSM1599037 COSM1117487 |
38 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782381476 | 40 | P>H | No |
ExAC gnomAD |
|
| rs2091865430 | 42 | T>A | No | Ensembl | |
| rs781955125 | 43 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs2091865488 | 44 | A>V | No | Ensembl | |
| rs782730265 | 45 | H>L | No | ExAC | |
| rs782730265 | 45 | H>P | No | ExAC | |
| rs782730265 | 45 | H>R | No | ExAC | |
|
rs1557059438 COSM1117488 COSM1599036 |
46 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 46 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1394011480 | 48 | S>P | No |
TOPMed gnomAD |
|
| rs782170598 | 49 | A>T | No |
ExAC gnomAD |
|
| rs782764739 | 52 | T>I | No |
ExAC gnomAD |
|
|
COSM355754 rs991662331 |
54 | L>F | lung [Cosmic] | No |
cosmic curated TOPMed |
|
COSM6186137 COSM6186138 |
60 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1233346105 | 61 | P>L | No |
TOPMed gnomAD |
|
| rs1435797206 | 62 | G>D | No |
TOPMed gnomAD |
|
| rs1203576071 | 62 | G>S | No |
TOPMed gnomAD |
|
| rs1557059469 | 63 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs202183031 | 63 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1557059474 | 66 | L>V | No |
TOPMed gnomAD |
|
| rs950261484 | 67 | Y>C | No | gnomAD | |
| rs950261484 | 67 | Y>F | No | gnomAD | |
| rs2091866301 | 69 | G>D | No | Ensembl | |
| rs781795736 | 70 | A>S | No |
ExAC TOPMed |
|
| rs781795736 | 70 | A>T | No |
ExAC TOPMed |
|
| rs782292845 | 73 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs782498132 | 73 | R>H | No |
ExAC TOPMed gnomAD |
|
|
rs782570670 COSM5219431 COSM1490647 |
75 | F>L | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC TOPMed gnomAD |
|
COSM3759411 COSM3759412 |
76 | Q>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782212030 | 77 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs139389280 | 80 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs781991732 | 84 | E>K | No |
ExAC gnomAD |
|
| rs781991732 | 84 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs782735815 | 85 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 85 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs147141867 | 86 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091866955 | 87 | A>P | No | Ensembl | |
| rs1486141388 | 92 | V>A | No |
TOPMed gnomAD |
|
| rs1208829849 | 94 | D>N | No |
TOPMed gnomAD |
|
| rs1557059503 | 97 | D>A | No |
TOPMed gnomAD |
|
| rs139449312 | 97 | D>E | No | ESP | |
| rs1557059503 | 97 | D>G | No |
TOPMed gnomAD |
|
| rs201281829 | 99 | V>M | No | TOPMed | |
| rs1223770494 | 100 | P>S | No | TOPMed | |
| rs144513627 | 102 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs148423464 | 102 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1354451731 | 105 | A>V | No |
TOPMed gnomAD |
|
| rs373948980 | 106 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091867466 | 107 | C>Y | No | TOPMed | |
|
rs1557059521 COSM4825342 COSM4825341 |
109 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| TCGA novel | 111 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557059525 | 113 | T>N | No | gnomAD | |
| rs2091867642 | 120 | L>R | No | Ensembl | |
| rs2091867612 | 120 | L>V | No | Ensembl | |
| rs2091867744 | 122 | V>L | No | TOPMed | |
| rs1557059533 | 123 | S>N | No | gnomAD | |
| rs1398924090 | 125 | R>C | No |
TOPMed gnomAD |
|
| rs368428119 | 125 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
|
rs34360382 VAR_050601 |
126 | A>T | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1257078950 | 127 | Q>E | No | TOPMed | |
| rs1741385319 | 130 | V>G | No | TOPMed | |
| rs781962004 | 130 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1032977318 | 132 | C>* | No |
TOPMed gnomAD |
|
| rs1215118708 | 133 | G>R | No | TOPMed | |
| rs1275542552 | 134 | Q>E | No |
TOPMed gnomAD |
|
| rs782237640 | 135 | V>A | No |
ExAC gnomAD |
|
| rs371894136 | 136 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs187773877 | 136 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1291804878 | 138 | G>S | No |
TOPMed gnomAD |
|
| rs1603241513 | 139 | V>A | No | TOPMed | |
| rs1603241513 | 139 | V>G | No | TOPMed | |
| rs2091868537 | 139 | V>M | No | TOPMed | |
| rs781908961 | 140 | C>* | No | ExAC | |
| rs2091868624 | 140 | C>R | No | TOPMed | |
| rs1427975271 | 141 | E>D | No | TOPMed | |
| rs1396609101 | 143 | R>P | No |
TOPMed gnomAD |
|
| rs1396609101 | 143 | R>Q | No |
TOPMed gnomAD |
|
| rs782794914 | 143 | R>W | No |
ExAC gnomAD |
|
| rs375561432 | 144 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs782121876 | 144 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs375561432 | 144 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1172246548 | 146 | G>E | No |
TOPMed gnomAD |
|
| rs1557059571 | 146 | G>R | No | gnomAD | |
| rs371848444 | 150 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs781839611 | 150 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs781839611 | 150 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs782715395 | 151 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs782715395 | 151 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs781908260 | 153 | Y>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 157 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs971617999 | 157 | D>H | No | Ensembl | |
| rs781825936 | 158 | P>S | No |
ExAC TOPMed gnomAD |
|
|
COSM3559515 COSM3559516 rs782575354 |
159 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs782484859 | 159 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs782484859 | 159 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1214922648 | 161 | G>R | No |
TOPMed gnomAD |
|
| rs1557059599 | 161 | G>V | No |
TOPMed gnomAD |
|
| rs2148413440 | 162 | Q>H | No | Ensembl | |
| rs2091869599 | 166 | A>P | No | TOPMed | |
| rs1264091848 | 169 | P>L | No |
TOPMed gnomAD |
|
| rs1264091848 | 169 | P>Q | No |
TOPMed gnomAD |
|
| rs2091869783 | 170 | G>R | No | TOPMed | |
| rs2148413486 | 172 | A>S | No | Ensembl | |
| rs1557059613 | 173 | T>M | No | gnomAD | |
| rs1212295165 | 174 | V>A | No |
TOPMed gnomAD |
|
| rs2091869987 | 176 | L>P | No | Ensembl | |
| rs1557059621 | 178 | V>A | No | gnomAD | |
| rs1440146266 | 179 | P>L | No |
TOPMed gnomAD |
|
| rs1557059626 | 181 | P>L | No | gnomAD | |
| rs868950741 | 182 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1355351960 | 182 | G>V | No | TOPMed | |
| rs781934370 | 183 | R>Q | No |
ExAC gnomAD |
|
| rs1170093749 | 183 | R>W | No |
TOPMed gnomAD |
|
| rs1557059637 | 184 | D>E | No | gnomAD | |
| rs2091870386 | 187 | L>F | No | TOPMed | |
|
COSM4646869 COSM1466593 rs369077780 |
193 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs782084008 | 195 | K>Q | No | ExAC | |
| rs1179329749 | 196 | L>P | No |
TOPMed gnomAD |
|
|
COSM3694421 rs782765277 COSM3694422 |
197 | S>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1349575283 | 199 | G>E | No | TOPMed | |
| rs1202661698 | 199 | G>W | No | Ensembl | |
| rs1557059664 | 200 | V>G | No | gnomAD | |
|
COSM3559517 COSM3559518 |
202 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4917266 COSM4917267 |
204 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2091870893 | 206 | R>C | No | Ensembl | |
| rs1557059668 | 206 | R>H | No | gnomAD | |
| rs2091870952 | 209 | A>T | No | TOPMed | |
| rs1308989605 | 210 | G>A | No | Ensembl | |
| rs781896492 | 210 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1308989605 | 210 | G>V | No | Ensembl | |
| rs781896492 | 210 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs150396453 | 212 | Q>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1557059684 | 215 | S>C | No | gnomAD | |
| rs1557059686 | 216 | S>G | No | gnomAD | |
| rs1372422105 | 216 | S>T | No | TOPMed | |
| rs2091871389 | 217 | E>D | No | Ensembl | |
|
COSM5135781 rs377546863 COSM1466594 |
217 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2091871429 | 218 | G>A | No | Ensembl | |
| rs369746764 | 219 | L>P | No |
ESP TOPMed gnomAD |
|
| rs149161688 | 221 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs782405462 | 221 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1434118229 | 223 | V>L | No | TOPMed | |
| rs1557059699 | 224 | V>L | No | gnomAD | |
| rs1557059707 | 226 | D>N | No | gnomAD | |
| rs373995578 | 229 | D>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs373995578 | 229 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1557059719 | 231 | N>D | No | gnomAD | |
| rs781970994 | 232 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs377510384 | 233 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs782025100 | 235 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs782468149 | 236 | G>A | No |
ExAC gnomAD |
|
| rs782754359 | 236 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs781787695 | 238 | F>L | No |
ExAC gnomAD |
|
| rs1569541698 | 239 | A>G | No | Ensembl | |
| rs782464330 | 239 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1313637681 | 240 | D>A | No | Ensembl | |
|
COSM3424604 rs782463555 COSM3424605 |
240 | D>N | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs370988850 | 241 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs370988850 | 241 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs200638079 | 241 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200638079 | 241 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs370988850 | 241 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs781945952 | 242 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs782281624 | 242 | R>H | No |
ExAC gnomAD |
|
| rs781975356 | 244 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2091872561 | 246 | F>I | No | Ensembl | |
| rs1418129228 | 246 | F>L | No |
TOPMed gnomAD |
|
| rs2091872561 | 246 | F>L | No | Ensembl | |
| rs782126578 | 247 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs782723671 | 249 | R>C | No |
ExAC gnomAD |
|
| rs374406078 | 249 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1205162516 | 250 | R>C | No |
TOPMed gnomAD |
|
| rs782047906 | 250 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1205162516 | 250 | R>S | No |
TOPMed gnomAD |
|
| rs782704025 | 251 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs781887159 | 251 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs781887159 | 251 | R>L | No |
ExAC gnomAD |
|
| rs782793837 | 252 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs781827670 | 254 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs199917520 | 254 | R>Q | No |
1000Genomes ExAC gnomAD |
|
| rs781827670 | 254 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs782552912 | 256 | Q>R | No |
ExAC gnomAD |
|
| rs1557059784 | 257 | A>P | No | gnomAD | |
| TCGA novel | 257 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782250509 | 260 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs376257187 | 260 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs376257187 | 260 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs781948973 | 262 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs1384892117 | 263 | V>M | No |
TOPMed gnomAD |
|
|
COSM3406111 COSM3406110 |
265 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1557059801 | 265 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs868921229 | 265 | R>S | No | gnomAD | |
| rs2091873504 | 266 | V>I | No | gnomAD | |
| rs2091873632 | 269 | G>E | No | TOPMed | |
| rs781933765 | 270 | D>N | No |
ExAC gnomAD |
|
| rs2091873674 | 271 | T>I | No | Ensembl | |
| rs909462323 | 277 | V>L | No | TOPMed | |
| rs909462323 | 277 | V>M | No | TOPMed | |
| rs370095364 | 279 | V>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs370095364 | 279 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1557059812 | 281 | L>F | No | gnomAD | |
| rs1557059812 | 281 | L>V | No | gnomAD | |
| rs782082970 | 282 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs782082970 | 282 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1427336226 | 283 | C>S | No | TOPMed | |
| rs2091874005 | 284 | Q>R | No | TOPMed | |
| rs374111928 | 285 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091874058 | 286 | Q>L | No | Ensembl | |
| rs1272367250 | 287 | G>D | No |
TOPMed gnomAD |
|
| rs1468845702 | 287 | G>S | No | TOPMed | |
| rs11798842 | 288 | L>F | No | Ensembl | |
| rs782073075 | 289 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs781911980 | 292 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1557059838 | 292 | A>V | No | gnomAD | |
| rs1354538665 | 295 | A>T | No |
TOPMed gnomAD |
|
| rs782503637 | 296 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs782503637 | 296 | P>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 300 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091874542 | 304 | A>T | No | Ensembl | |
| rs2148414095 | 304 | A>V | No | Ensembl | |
| rs1443804810 | 305 | A>P | No |
TOPMed gnomAD |
|
| rs1443804810 | 305 | A>T | No |
TOPMed gnomAD |
|
| rs782352020 | 305 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1403895895 | 306 | G>S | No | TOPMed | |
| TCGA novel | 307 | P>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091874797 | 307 | P>R | No | TOPMed | |
| rs782338582 | 309 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs781972411 | 310 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs202085462 | 312 | A>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2091874907 | 312 | A>P | No | TOPMed | |
|
rs202085462 COSM1117491 COSM1599033 |
312 | A>V | endometrium [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1454684044 | 313 | A>V | No |
TOPMed gnomAD |
|
| rs1557059889 | 316 | A>P | No | gnomAD | |
|
COSM1715847 COSM1715848 |
318 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1557059894 | 318 | P>S | No |
TOPMed gnomAD |
|
| rs1557059897 | 319 | M>I | No | gnomAD | |
| rs1557059896 | 319 | M>V | No |
TOPMed gnomAD |
|
| rs2091875288 | 320 | V>A | No | Ensembl | |
| rs1439273120 | 320 | V>M | No |
TOPMed gnomAD |
|
| rs2091875313 | 321 | E>K | No | TOPMed | |
| rs2091875375 | 323 | G>S | No | Ensembl | |
| rs781868142 | 324 | A>T | No |
ExAC gnomAD |
|
| rs2091875468 | 325 | S>G | No | TOPMed | |
| TCGA novel | 325 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374183010 | 326 | M>I | No |
ESP ExAC gnomAD |
|
| rs2091875526 | 329 | A>V | No | Ensembl | |
| rs1341698175 | 330 | R>Q | No |
TOPMed gnomAD |
|
| rs1557059904 | 330 | R>W | No | gnomAD | |
| rs2148414218 | 331 | R>I | No | Ensembl | |
| rs1557059910 | 335 | T>A | No | gnomAD | |
| rs1236522993 | 335 | T>M | No |
TOPMed gnomAD |
|
| rs2091875818 | 336 | A>T | No | Ensembl | |
| rs782669923 | 336 | A>V | No |
ExAC TOPMed gnomAD |
|
|
rs1557059915 CA415097737 RCV000498811 |
337 | G>R | No |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
| rs1337959515 | 338 | G>C | No |
TOPMed gnomAD |
|
| rs2091876032 | 338 | G>D | No | Ensembl | |
| rs1337959515 | 338 | G>S | No |
TOPMed gnomAD |
|
| rs782600752 | 339 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs782600752 | 339 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs782506512 | 339 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2091876215 | 340 | G>D | No | TOPMed | |
| rs782787744 | 340 | G>S | No |
1000Genomes gnomAD |
|
| rs2091876272 | 341 | P>T | No | Ensembl | |
| rs1175001921 | 342 | S>G | No | TOPMed | |
| rs782305118 | 342 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs1434738521 | 343 | G>C | No |
TOPMed gnomAD |
|
| rs1434738521 | 343 | G>S | No |
TOPMed gnomAD |
|
| rs1170819184 | 344 | A>T | No |
TOPMed gnomAD |
|
| rs2091876396 | 345 | E>K | No | Ensembl | |
| rs1233293552 | 346 | E>D | No | TOPMed | |
| rs1471112825 | 346 | E>V | No |
TOPMed gnomAD |
|
| rs2091876482 | 347 | A>S | No | Ensembl | |
| rs1557059939 | 349 | V>M | No |
TOPMed gnomAD |
|
| rs1557059940 | 350 | E>G | No | gnomAD | |
| rs2091876648 | 352 | G>A | No | Ensembl | |
| rs368901623 | 352 | G>S | No |
ESP TOPMed gnomAD |
|
| rs2091876741 | 353 | V>A | No | TOPMed | |
| rs782055449 | 353 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs868988904 | 354 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1006325245 | 356 | R>C | No |
TOPMed gnomAD |
|
| rs1280542430 | 356 | R>H | No |
TOPMed gnomAD |
|
| rs1409552761 | 357 | C>R | No |
TOPMed gnomAD |
|
| rs1314259821 | 358 | V>I | No |
TOPMed gnomAD |
|
| rs1415015783 | 359 | T>I | No |
TOPMed gnomAD |
|
| rs2091877106 | 361 | P>S | No | TOPMed | |
| rs782488240 | 363 | D>A | No | ExAC | |
| rs1365303923 | 363 | D>E | No |
TOPMed gnomAD |
|
| rs868907205 | 364 | S>F | No |
TOPMed gnomAD |
|
| rs868907205 | 364 | S>Y | No |
TOPMed gnomAD |
|
| rs2091882247 | 365 | P>S | No | TOPMed | |
| TCGA novel | 366 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782524207 | 366 | E>G | No |
ExAC gnomAD |
|
| rs781800887 | 366 | E>K | No |
ExAC TOPMed gnomAD |
|
|
COSM3559520 COSM3559521 rs1444686074 COSM3559519 |
367 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1168063118 | 368 | Y>* | No |
TOPMed gnomAD |
|
| rs1557060093 | 369 | P>T | No | gnomAD | |
| rs782511958 | 372 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2091882482 | 373 | E>K | No | TOPMed | |
| rs1031438719 | 374 | H>D | No |
TOPMed gnomAD |
|
| rs782239104 | 375 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs782239104 | 375 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs2091882516 | 375 | T>P | No | Ensembl | |
| rs782239104 | 375 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs2148415159 | 377 | S>R | No | Ensembl | |
| rs1433090818 | 378 | P>T | No | gnomAD | |
| rs1557060102 | 379 | I>V | No | gnomAD | |
| rs782316937 | 381 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs782278304 | 382 | R>C | No |
ExAC gnomAD |
|
| rs782418950 | 382 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1603242347 | 384 | P>A | No | TOPMed | |
| rs2091882772 | 384 | P>H | No | Ensembl | |
| rs1603242347 | 384 | P>S | No | TOPMed | |
| rs1557060117 | 385 | L>Q | No | gnomAD | |
| rs782134831 | 386 | E>D | No |
ExAC gnomAD |
|
| rs2148415199 | 389 | P>H | No | Ensembl | |
| rs781919430 | 390 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs782791284 | 391 | L>R | No |
ExAC gnomAD |
|
| rs782507197 | 392 | K>M | No |
ExAC gnomAD |
|
| rs782764110 | 392 | K>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 393 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091882965 | 394 | G>E | No | TOPMed | |
| rs782575059 | 394 | G>R | No |
ExAC gnomAD |
|
| rs782203925 | 395 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs782540845 | 396 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs141165767 | 397 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1557060146 | 398 | S>N | No |
TOPMed gnomAD |
|
| rs1408216858 | 400 | V>A | No | TOPMed | |
| rs371643094 | 400 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs371643094 | 400 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs782017806 | 406 | D>E | No |
ExAC gnomAD |
|
|
COSM322729 rs2091883295 |
406 | D>N | lung [Cosmic] | No |
cosmic curated gnomAD |
|
COSM1292957 rs376853068 |
407 | G>R | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2091883394 | 408 | H>N | No | TOPMed | |
| rs1466003030 | 409 | M>T | No | TOPMed | |
| rs781997206 | 410 | I>L | No |
ExAC gnomAD |
|
| rs782073875 | 410 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs782753066 | 415 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs201740104 | 416 | T>I | No |
1000Genomes ExAC |
|
| rs201740104 | 416 | T>S | No |
1000Genomes ExAC |
|
| rs1557060198 | 417 | Q>R | No |
TOPMed gnomAD |
|
| rs1557060205 | 419 | Q>R | No | gnomAD | |
| rs1473086829 | 420 | L>V | No |
TOPMed gnomAD |
|
| rs1557060216 | 421 | Y>* | No | Ensembl | |
| rs1557060214 | 421 | Y>C | No | gnomAD | |
| rs1557060220 | 422 | K>N | No |
TOPMed gnomAD |
|
| rs1413220700 | 426 | H>P | No | TOPMed | |
| rs782281721 | 427 | G>D | No |
ExAC gnomAD |
|
| rs1557060480 | 427 | G>S | No |
TOPMed gnomAD |
|
| rs782281721 | 427 | G>V | No |
ExAC gnomAD |
|
| TCGA novel | 428 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782438257 | 429 | Q>R | No |
ExAC gnomAD |
|
| rs1557060486 | 431 | Q>H | No |
TOPMed gnomAD |
|
| rs2091891920 | 431 | Q>R | No | TOPMed | |
| rs973814086 | 432 | V>I | No | Ensembl | |
| rs2091891977 | 433 | Y>H | No | Ensembl | |
| rs1451908302 | 434 | H>Y | No | TOPMed | |
| rs782347144 | 437 | Q>K | No |
ExAC gnomAD |
|
| rs2091892059 | 440 | P>T | No | TOPMed | |
| rs781965537 | 441 | P>A | No |
ExAC gnomAD |
|
|
COSM1599032 COSM1117492 |
441 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782051376 | 442 | G>D | No |
ExAC gnomAD |
|
| rs2091892130 | 443 | S>L | No |
TOPMed gnomAD |
|
| rs139115534 | 444 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1221914442 | 445 | I>V | No | TOPMed | |
| rs782772907 | 447 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs782772907 | 447 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs781819769 | 450 | L>Q | No |
ExAC gnomAD |
|
| rs953495911 | 451 | L>M | No | Ensembl | |
| rs144016495 | 451 | L>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs146005579 | 452 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1229892136 | 452 | D>G | No | TOPMed | |
|
COSM4920973 COSM4920974 |
452 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs139900515 | 453 | S>N | No |
ESP TOPMed gnomAD |
|
| rs1432789842 | 454 | S>N | No | TOPMed | |
| TCGA novel | 455 | G>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782555071 | 455 | G>A | No |
ExAC gnomAD |
|
| rs782679643 | 456 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs782679643 | 456 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1369524419 | 457 | H>D | No |
TOPMed gnomAD |
|
| rs782248987 | 457 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1369524419 | 457 | H>Y | No |
TOPMed gnomAD |
|
| rs2091892605 | 459 | Y>H | No | Ensembl | |
| rs199597733 | 462 | T>A | No |
1000Genomes ExAC gnomAD |
|
| rs988278268 | 463 | A>T | No |
TOPMed gnomAD |
|
| rs2091892690 | 463 | A>V | No | TOPMed | |
| rs1569541790 | 464 | H>P | No | Ensembl | |
| rs2091892729 | 464 | H>Y | No | Ensembl | |
| rs782323500 | 467 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs782174999 | 468 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs782554732 COSM1117493 COSM1651540 |
468 | R>W | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1557060605 | 469 | I>M | No | gnomAD | |
| rs1183959770 | 470 | P>L | No | TOPMed | |
| rs782380699 | 470 | P>S | No | ExAC | |
| rs782083217 | 473 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs782083217 | 473 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs376423175 | 475 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1557060611 | 475 | P>T | No | gnomAD | |
| rs1484203168 | 476 | Q>* | No |
TOPMed gnomAD |
|
| rs1036115653 | 476 | Q>H | No |
TOPMed gnomAD |
|
| rs1484203168 | 476 | Q>K | No |
TOPMed gnomAD |
|
| rs2148416913 | 476 | Q>R | No | Ensembl | |
| rs782809682 | 478 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs782809682 | 478 | P>R | No |
ExAC gnomAD |
|
| rs1557060618 | 479 | D>H | No |
TOPMed gnomAD |
|
| rs1557060618 | 479 | D>Y | No |
TOPMed gnomAD |
|
| rs1603243301 | 480 | C>Y | No | Ensembl | |
| rs1557060624 | 482 | S>R | No | gnomAD | |
| rs2091895049 | 483 | C>F | No | Ensembl | |
| rs28447247 | 487 | Q>R | No | Ensembl | |
| rs201696853 | 489 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1557060629 | 489 | P>T | No | gnomAD | |
| rs1557060636 | 490 | L>P | No | gnomAD | |
| rs782445788 | 493 | W>L | No |
ExAC gnomAD |
|
| rs1557060640 | 494 | C>G | No | Ensembl | |
| rs2091895368 | 499 | R>K | No | TOPMed | |
|
COSM6118146 COSM6118147 COSM6118148 |
500 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782275965 | 502 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs782275965 | 502 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs782453850 | 502 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs782367079 | 503 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs782001393 | 504 | G>D | No |
ExAC gnomAD |
|
|
RCV000964158 rs145651760 |
507 | G>R | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs145651760 | 507 | G>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782803644 | 508 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs782060934 | 508 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs542027507 | 510 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs148901418 | 510 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs781830850 | 511 | Q>H | No |
ExAC gnomAD |
|
| rs782496204 | 512 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs781809734 | 517 | W>C | No |
ExAC gnomAD |
|
| rs79210580 | 517 | W>G | No |
ExAC gnomAD |
|
| rs782531618 | 518 | S>G | No |
ExAC gnomAD |
|
| rs782692577 | 518 | S>R | No | ExAC | |
| rs782255143 | 519 | Y>F | No | ExAC | |
| rs1557060936 | 521 | E>D | No | gnomAD | |
| rs2091907017 | 521 | E>K | No | gnomAD | |
| rs1557060941 | 522 | D>A | No | gnomAD | |
| rs1557060939 | 522 | D>N | No | gnomAD | |
| rs2091907089 | 523 | S>G | No | TOPMed | |
| rs782477518 | 523 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs782618067 | 524 | H>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
|
COSM1466597 COSM5094229 COSM5094230 |
525 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782013908 | 528 | I>S | No |
ExAC gnomAD |
|
| rs782013908 | 528 | I>T | No |
ExAC gnomAD |
|
| rs2091907202 | 529 | Q>H | No | TOPMed | |
| rs2091907222 | 530 | S>N | No | TOPMed | |
| rs2091907242 | 531 | L>M | No | TOPMed | |
| rs782284503 | 533 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1557060959 | 534 | G>S | No | gnomAD | |
| rs2148418818 | 536 | H>N | No | Ensembl | |
| rs2091907359 | 536 | H>P | No | Ensembl | |
| rs1557060961 | 536 | H>Q | No | gnomAD | |
| TCGA novel | 538 | R>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs370097760 | 538 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs370097760 | 538 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs782808628 | 538 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs782808628 | 538 | R>P | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 540 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs781836984 RCV000943132 |
540 | E>K | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| COSM422193 | 541 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2091907501 | 541 | Q>R | No | TOPMed | |
| rs782429021 | 542 | G>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1243554267 | 542 | G>D | No | TOPMed | |
| rs782429021 | 542 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781838386 | 543 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs782718017 | 543 | Q>R | No |
ExAC gnomAD |
|
| TCGA novel | 544 | V>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557061068 | 544 | V>I | No | gnomAD | |
| rs1557061068 | 544 | V>L | No | gnomAD | |
| rs782438501 | 545 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs782438501 | 545 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs782595560 | 546 | L>F | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 547 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782426429 | 548 | V>A | No |
ExAC gnomAD |
|
| rs938008258 | 549 | P>L | No | Ensembl | |
| rs2091910691 | 549 | P>S | No | TOPMed | |
| rs2091910691 | 549 | P>T | No | TOPMed | |
|
COSM3732432 COSM3732431 COSM3732430 |
550 | R>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs782213788 VAR_079495 |
550 | R>Q | No |
UniProt ExAC TOPMed dbSNP gnomAD |
|
| rs782636433 | 550 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2091910825 | 553 | I>N | No | Ensembl | |
| rs1557061091 | 555 | D>H | No | gnomAD | |
| rs1557061091 | 555 | D>N | No | gnomAD | |
| rs2091910904 | 556 | A>E | No | Ensembl | |
| rs1055646249 | 557 | D>H | No |
TOPMed gnomAD |
|
| rs1055646249 | 557 | D>N | No |
TOPMed gnomAD |
|
|
COSM1599031 COSM1117494 |
558 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781918516 | 558 | E>K | No |
ExAC gnomAD |
|
| rs782394886 | 560 | F>L | No |
ExAC gnomAD |
|
| rs782112812 | 563 | A>V | No |
ExAC gnomAD |
|
| rs2266878 | 565 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC TOPMed gnomAD NCI-TCGA |
| rs782708590 | 566 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs782708590 | 566 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1557061121 | 568 | D>N | No |
TOPMed gnomAD |
|
| rs1557061121 | 568 | D>Y | No |
TOPMed gnomAD |
|
| rs1490875008 | 569 | S>T | No | TOPMed | |
| rs1557061123 | 570 | L>F | No | gnomAD | |
|
COSM1599030 COSM1117495 |
572 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1037119823 | 572 | H>Q | No | Ensembl | |
| rs781812882 | 573 | V>M | No |
ExAC gnomAD |
|
| rs1557061131 | 575 | G>E | No | gnomAD | |
| rs1222442058 | 575 | G>R | No | TOPMed | |
| rs1222442058 | 575 | G>W | No | TOPMed | |
| rs782548296 | 577 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs782366597 | 577 | H>Y | No | 1000Genomes | |
|
COSM1726724 rs782803101 COSM1726723 |
578 | V>M | liver [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1557061140 | 579 | A>S | No | gnomAD | |
| rs781858832 | 580 | C>S | No |
ExAC gnomAD |
|
| rs1228530572 | 581 | V>I | No | TOPMed | |
| rs1326206620 | 583 | P>S | No |
TOPMed gnomAD |
|
| rs1557061150 | 584 | P>S | No | gnomAD | |
| rs782614928 | 585 | Q>P | No |
ExAC gnomAD |
|
| rs146501701 | 586 | D>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1603244549 | 587 | Q>H | No | Ensembl | |
| rs2091911546 | 588 | V>A | No | Ensembl | |
| rs782455705 | 591 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs1365707973 | 591 | N>T | No |
TOPMed gnomAD |
|
| rs1557061158 | 592 | P>A | No | gnomAD | |
| rs1557061158 | 592 | P>S | No | gnomAD | |
| rs782283893 | 593 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs2091911648 | 593 | P>Q | No | Ensembl | |
|
COSM3559523 COSM3559524 COSM3559522 |
593 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782372087 | 594 | G>S | No |
ExAC gnomAD |
|
| rs2091913697 | 596 | D>A | No | Ensembl | |
|
COSM1715849 COSM1715850 |
597 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2091913783 | 598 | V>A | No | Ensembl | |
|
rs2266879 VAR_019681 |
598 | V>I | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1557061244 | 600 | V>M | No | gnomAD | |
| rs200197753 | 601 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782812513 | 604 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs949435109 | 605 | M>I | No | TOPMed | |
| rs2091913956 | 605 | M>T | No | TOPMed | |
| rs1557061252 | 606 | F>L | No |
TOPMed gnomAD |
|
| rs374123296 | 607 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs367854471 | 609 | V>M | No |
ESP TOPMed gnomAD |
|
| rs995669353 | 611 | V>L | No | Ensembl | |
| rs2091914125 | 612 | A>T | No | TOPMed | |
| TCGA novel | 613 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 617 | S>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091914168 | 620 | D>V | No | Ensembl | |
| rs782552714 | 622 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs375109307 | 623 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 623 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1117498 COSM1651538 rs539782168 COSM4378301 |
624 | V>I | Variant assessed as Somatic; MODERATE impact. endometrium haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs782576131 | 625 | Q>P | No |
ExAC gnomAD |
|
| rs782190621 | 627 | L>F | No |
ExAC gnomAD |
|
| rs2148419922 | 627 | L>M | No | Ensembl | |
| rs782417459 | 628 | E>K | No |
ExAC TOPMed gnomAD |
|
|
COSM4911228 rs782036174 COSM4911229 COSM4911227 |
629 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC TOPMed gnomAD |
| rs1338462583 | 629 | A>T | No |
TOPMed gnomAD |
|
| rs782036174 | 629 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1336380813 | 630 | A>T | No | Ensembl | |
| rs2091914459 | 631 | A>D | No | Ensembl | |
| rs781950083 | 631 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs781950083 | 631 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs372331714 | 632 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs372331714 | 632 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs782775543 | 634 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs781861323 | 634 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs147082716 | 635 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1603244975 | 636 | C>R | No | Ensembl | |
| rs782461403 | 637 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs782002708 | 638 | G>A | No | ExAC | |
| rs782377121 | 638 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs782002708 | 638 | G>V | No | ExAC | |
| TCGA novel | 639 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782209764 | 641 | W>S | No |
ExAC gnomAD |
|
| rs782469938 | 642 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1289345640 | 642 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2091916506 | 644 | H>N | No | Ensembl | |
| rs781926148 | 647 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs781926148 | 647 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs782809651 | 648 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs1338558224 | 648 | Q>H | No |
TOPMed gnomAD |
|
| rs781983227 | 649 | S>I | No |
ExAC gnomAD |
|
| rs190195934 | 649 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2091916670 | 650 | S>G | No | TOPMed | |
| rs1557061385 | 651 | H>R | No |
TOPMed gnomAD |
|
| rs781832307 | 653 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs181891775 | 654 | Y>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782434553 | 654 | Y>H | No |
ExAC gnomAD |
|
| rs181891775 | 654 | Y>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1412060428 | 655 | G>A | No |
TOPMed gnomAD |
|
|
rs782559344 COSM5090365 COSM1466600 COSM5090366 |
655 | G>R | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1557061397 | 656 | E>K | No | gnomAD | |
| rs1557061397 | 656 | E>Q | No | gnomAD | |
| rs1557061403 | 659 | P>R | No | gnomAD | |
| rs1557061408 | 660 | E>A | No | gnomAD | |
| rs1185620017 | 660 | E>K | No | TOPMed | |
| rs1185620017 | 660 | E>Q | No | TOPMed | |
| rs1270448175 | 661 | G>D | No |
TOPMed gnomAD |
|
| rs1476606812 | 661 | G>S | No |
TOPMed gnomAD |
|
| rs373196958 | 662 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091917033 | 663 | R>G | No | TOPMed | |
| rs782481792 | 663 | R>M | No |
ExAC TOPMed gnomAD |
|
| rs782186077 | 663 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs782481792 | 663 | R>T | No |
ExAC TOPMed gnomAD |
|
|
RCV000879484 rs147973343 |
664 | T>I | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2091917107 | 664 | T>P | No | TOPMed | |
| rs145979763 | 668 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs145979763 | 668 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs782305028 | 668 | A>V | No |
ExAC gnomAD |
|
| rs782153200 | 670 | E>D | No |
ExAC TOPMed gnomAD |
|
|
rs139853034 RCV000885932 |
670 | E>K | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1569541878 | 671 | V>A | No | TOPMed | |
| rs1557061507 | 671 | V>L | No |
TOPMed gnomAD |
|
| rs1557061507 | 671 | V>M | No |
TOPMed gnomAD |
|
| rs895001229 | 672 | D>Y | No | Ensembl | |
| rs199908902 | 674 | Q>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782294628 | 675 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs782294628 | 675 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs150768754 | 675 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM3694204 COSM3694205 COSM3694203 rs782515421 |
676 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1165975 rs139203705 |
676 | R>H | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs782515421 | 676 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1557061520 | 679 | G>E | No | gnomAD | |
| rs2091919938 | 679 | G>R | No | Ensembl | |
| rs2091920001 | 680 | A>D | No | gnomAD | |
| rs782215577 | 680 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs782215577 | 680 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2091920021 | 681 | C>G | No | Ensembl | |
| rs781975103 | 683 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC TOPMed gnomAD NCI-TCGA |
| rs2091920076 | 684 | V>I | No | Ensembl | |
| rs781982131 | 685 | E>G | No |
ExAC gnomAD |
|
| rs782058864 | 685 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782121909 | 687 | L>Q | No |
ExAC gnomAD |
|
| rs2094675809 | 688 | A>T | No | Ensembl | |
| rs1557061530 | 688 | A>V | No | gnomAD | |
| rs782738493 | 689 | G>D | No |
ExAC gnomAD |
|
| rs2091920176 | 689 | G>R | No | TOPMed | |
| rs1025679740 | 690 | P>L | No | TOPMed | |
| rs2091920284 | 692 | L>V | No | TOPMed | |
| rs2091920315 | 693 | V>L | No | Ensembl | |
| rs782715367 | 695 | V>M | No |
ExAC gnomAD |
|
| rs2148420786 | 696 | G>R | No | Ensembl | |
| rs782757941 | 697 | W>* | No |
ExAC gnomAD |
|
| rs782560262 | 697 | W>S | No |
ExAC TOPMed gnomAD |
|
| rs145207517 | 698 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1238586993 | 699 | S>N | No |
TOPMed gnomAD |
|
| rs1557061544 | 700 | H>Y | No | gnomAD | |
|
COSM3747367 COSM3747368 rs1569541885 |
702 | A>V | liver [Cosmic] | No |
cosmic curated Ensembl |
|
TCGA novel rs1557061551 |
704 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
rs373965130 COSM5711540 COSM5711541 COSM5711539 |
704 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP NCI-TCGA TOPMed gnomAD |
| rs1557061551 | 704 | R>S | No | gnomAD | |
| rs782548925 | 705 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782548925 | 705 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782236263 | 706 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs201817979 | 706 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM4107815 COSM4107814 rs782236263 COSM4107813 |
706 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1599028 COSM1117499 |
708 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781864901 | 710 | H>L | No |
1000Genomes ExAC gnomAD |
|
| rs781864901 | 710 | H>R | No |
1000Genomes ExAC gnomAD |
|
| rs1557061570 | 710 | H>Y | No | Ensembl | |
|
COSM6186132 COSM6186131 COSM6186133 |
711 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1453928876 | 712 | R>* | No | TOPMed | |
|
COSM1599027 COSM4589281 rs368450576 COSM1117500 |
712 | R>Q | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs781954635 | 713 | G>A | No |
ExAC gnomAD |
|
| rs781954635 | 713 | G>D | No |
ExAC gnomAD |
|
| rs2091922294 | 713 | G>S | No | TOPMed | |
| rs1683292188 | 716 | A>D | No | TOPMed | |
| rs782099950 | 716 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs782099950 | 716 | A>T | No |
ExAC TOPMed gnomAD |
|
|
COSM4107816 COSM4107818 COSM4107817 |
717 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 717 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs140681169 RCV000912212 |
719 | H>R | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1557061626 | 721 | W>* | No | gnomAD | |
| rs2091922496 | 721 | W>* | No | TOPMed | |
|
COSM755755 COSM1648253 |
721 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1557061624 | 721 | W>R | No |
TOPMed gnomAD |
|
| rs1557061626 | 721 | W>S | No | gnomAD | |
| rs782155105 | 723 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1366051110 | 724 | L>V | No | TOPMed | |
| rs1557061639 | 725 | P>H | No | gnomAD | |
| rs2091922608 | 726 | G>R | No | Ensembl | |
| TCGA novel | 727 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782080381 | 727 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1456256645 | 727 | E>Q | No |
TOPMed gnomAD |
|
| rs1195633753 | 728 | L>P | No |
TOPMed gnomAD |
|
| rs147911027 | 729 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3235936 rs144605528 COSM4378309 COSM3235935 |
729 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs782464387 | 730 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs782464387 | 730 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs2091922722 | 731 | L>P | No | TOPMed | |
| rs782656549 | 732 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1209734596 | 733 | A>T | No | TOPMed | |
| rs1557061655 | 734 | T>I | No | gnomAD | |
| rs782654734 | 735 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1253394027 | 737 | E>* | No | TOPMed | |
| rs1227746011 | 737 | E>D | No |
TOPMed gnomAD |
|
| rs1253394027 | 737 | E>K | No | TOPMed | |
| rs2091922969 | 739 | A>V | No |
TOPMed gnomAD |
|
| rs1557061667 | 740 | G>E | No | gnomAD | |
| rs782360864 | 740 | G>R | No |
ExAC gnomAD |
|
| rs782425712 | 741 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs782198887 | 741 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1557061677 | 744 | L>V | No | gnomAD | |
| TCGA novel | 745 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1603245520 | 746 | H>N | No | Ensembl | |
| rs782034676 | 748 | Q>R | No |
ExAC gnomAD |
|
| rs373921353 | 749 | A>G | No |
ESP ExAC gnomAD |
|
| rs781968139 | 750 | H>D | No |
ExAC gnomAD |
|
| rs781968139 | 750 | H>Y | No |
ExAC gnomAD |
|
| TCGA novel | 751 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781946256 | 753 | Y>H | No |
1000Genomes ExAC gnomAD |
|
| rs782052980 | 755 | S>P | No |
ExAC gnomAD |
|
| rs782717176 | 756 | M>I | No |
ExAC gnomAD |
|
| rs1018657753 | 756 | M>T | No | Ensembl | |
| rs1557061777 | 759 | R>G | No |
TOPMed gnomAD |
|
| rs964030825 | 759 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1557061777 | 759 | R>W | No |
TOPMed gnomAD |
|
| rs782552228 | 760 | E>Q | No |
ExAC gnomAD |
|
| rs782781381 | 763 | V>M | No |
ExAC gnomAD |
|
| rs1333735113 | 765 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2091927616 | 766 | Y>C | No | Ensembl | |
| rs782425266 | 767 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs782565404 | 768 | T>S | No |
ExAC gnomAD |
|
|
rs1557061795 RCV001573651 |
769 | Q>H | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs782126891 | 769 | Q>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1557061800 | 770 | G>D | No | gnomAD | |
| rs1159833717 | 770 | G>S | No | TOPMed | |
| rs782682163 | 772 | A>P | No |
ExAC gnomAD |
|
| rs782682163 | 772 | A>T | No |
ExAC gnomAD |
|
| rs2091927871 | 773 | Q>E | No | TOPMed | |
| rs1557061807 | 776 | D>H | No | gnomAD | |
| rs2091927952 | 777 | N>S | No | gnomAD | |
| rs977962667 | 778 | T>A | No | gnomAD | |
| rs2091928018 | 779 | H>P | No | Ensembl | |
| rs2091928018 | 779 | H>R | No | Ensembl | |
| rs1557061813 | 780 | A>T | No |
TOPMed gnomAD |
|
| rs1476166336 | 784 | I>F | No |
TOPMed gnomAD |
|
| rs1263277859 | 784 | I>T | No |
TOPMed gnomAD |
|
| rs1223091634 | 787 | D>E | No |
TOPMed gnomAD |
|
| rs141109198 | 787 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs141109198 | 787 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs782155380 | 789 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs781871704 | 790 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs782760643 | 790 | M>V | No |
ExAC gnomAD |
|
| rs782080603 | 791 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs138935872 RCV000970525 |
793 | P>L | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs782451122 | 798 | C>Y | No |
ExAC gnomAD |
|
| rs782674956 | 800 | A>P | No |
ExAC gnomAD |
|
| rs781903961 | 800 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2091930117 | 802 | N>S | No | TOPMed | |
| rs782658604 | 803 | R>K | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 803 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782220156 | 804 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs782306101 | 805 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1557061879 | 806 | G>R | No | gnomAD | |
|
COSM3800479 COSM3800480 COSM3800481 |
807 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782570237 | 810 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs1423968200 | 812 | D>G | No |
TOPMed gnomAD |
|
| rs1423968200 | 812 | D>V | No |
TOPMed gnomAD |
|
| rs144601801 | 813 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091930403 | 814 | Q>* | No | TOPMed | |
| rs782255019 | 815 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs782345456 | 818 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs781971591 | 818 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1603246041 | 819 | Y>H | No | Ensembl | |
| rs1557061906 | 820 | G>E | No | gnomAD | |
| rs1488080718 | 820 | G>R | No | TOPMed | |
| rs782053902 | 821 | P>H | No |
ExAC gnomAD |
|
| rs1244780835 | 821 | P>S | No | TOPMed | |
| rs782714649 | 822 | L>S | No |
ExAC gnomAD |
|
| rs370781491 | 824 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 824 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782760077 | 825 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 825 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091930661 | 826 | G>E | No | Ensembl | |
| rs782573626 | 827 | A>S | No |
TOPMed gnomAD |
|
| rs782573626 | 827 | A>T | No |
TOPMed gnomAD |
|
| TCGA novel | 827 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557061926 | 828 | V>M | No | gnomAD | |
|
rs375202688 COSM3992414 COSM3992413 COSM3992412 |
829 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2091930850 | 833 | P>H | No | Ensembl | |
| rs782686812 | 834 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1444897449 | 834 | A>V | No |
TOPMed gnomAD |
|
| rs1330316647 | 835 | P>S | No | TOPMed | |
|
rs782525744 COSM1117501 COSM1651537 |
836 | S>N | endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs879975545 | 836 | S>R | No | Ensembl | |
| rs1557061944 | 839 | A>V | No | gnomAD | |
| rs1423421078 | 841 | E>K | No |
TOPMed gnomAD |
|
| rs782232949 | 845 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs782232949 | 845 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs782232949 | 845 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
COSM5129015 COSM5129014 COSM1466602 |
846 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782311955 | 846 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1557062050 | 847 | P>L | No | gnomAD | |
| rs2091934687 | 847 | P>S | No | Ensembl | |
| rs782371490 | 852 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs782371490 | 852 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 852 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091934822 | 853 | L>V | No | TOPMed | |
| rs1488656820 | 855 | I>V | No |
TOPMed gnomAD |
|
| rs1557062063 | 857 | G>A | No | gnomAD | |
| rs2091934904 | 859 | N>S | No | Ensembl | |
| rs138618805 | 862 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs138618805 | 862 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs782757118 | 862 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2091935040 | 864 | F>S | No | TOPMed | |
| rs201993079 | 865 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782651975 | 866 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs781828875 | 868 | Q>L | No |
ExAC gnomAD |
|
| rs782028379 | 870 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1117502 rs782630918 COSM1599026 |
871 | V>M | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs144548620 | 872 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091935279 | 873 | V>M | No | gnomAD | |
|
rs2091935302 COSM1117503 COSM1651536 |
875 | S>N | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
|
COSM6030145 rs141796383 COSM6030144 COSM6030146 |
876 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs148448651 | 876 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs782320094 | 879 | N>D | No |
ExAC gnomAD |
|
| rs781951810 | 881 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1557062115 | 881 | E>Q | No | gnomAD | |
| rs1557062120 | 882 | P>S | No | gnomAD | |
| rs200750569 | 883 | S>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 885 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557062125 | 885 | Y>F | No | Ensembl | |
| rs1453766008 | 886 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs782764892 | 886 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1453766008 | 886 | R>S | No |
TOPMed gnomAD |
|
| rs781873944 | 887 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1557062131 | 888 | S>L | No | gnomAD | |
| rs1557062136 | 889 | A>T | No | gnomAD | |
| rs782796672 | 890 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs140590097 | 890 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs782796672 | 890 | R>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 891 | I>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782613407 | 892 | V>L | No |
ExAC gnomAD |
|
| rs782319488 | 894 | V>L | No |
ExAC gnomAD |
|
| rs782319488 | 894 | V>M | No |
ExAC gnomAD |
|
| rs2091938912 | 897 | P>R | No | Ensembl | |
| rs2091938969 | 899 | P>H | No | gnomAD | |
| rs2091938992 | 900 | N>D | No | Ensembl | |
| rs782289362 | 900 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1557062246 | 902 | T>S | No | gnomAD | |
| rs782368588 | 905 | P>L | No |
ExAC gnomAD |
|
| rs201702238 | 906 | V>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201702238 | 906 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200934815 | 907 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782733468 | 907 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs782065187 | 908 | V>L | No |
ExAC gnomAD |
|
| rs782065187 | 908 | V>M | No |
ExAC gnomAD |
|
| rs782794387 | 909 | A>D | No |
ExAC gnomAD |
|
| rs1557062256 | 909 | A>T | No | gnomAD | |
| rs782794387 | 909 | A>V | No |
ExAC gnomAD |
|
| rs2091939211 | 911 | K>N | No |
TOPMed gnomAD |
|
| rs1603246586 | 912 | S>N | No | Ensembl | |
| rs1351190011 | 914 | P>S | No |
TOPMed gnomAD |
|
| rs1351190011 | 914 | P>T | No |
TOPMed gnomAD |
|
| rs2091939289 | 915 | P>A | No | Ensembl | |
| rs1278049497 | 920 | Q>H | No |
TOPMed gnomAD |
|
| rs1450407349 | 922 | F>L | No | Ensembl | |
| rs1279464807 | 926 | D>A | No |
TOPMed gnomAD |
|
| rs1557062495 | 933 | S>N | No |
TOPMed gnomAD |
|
| rs1557062495 | 933 | S>T | No |
TOPMed gnomAD |
|
| TCGA novel | 934 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557062498 | 934 | P>T | No | gnomAD | |
| rs2148424746 | 935 | R>C | No | Ensembl | |
| rs370158582 | 935 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1603247121 | 938 | P>L | No | TOPMed | |
| rs868933178 | 939 | Q>K | No | TOPMed | |
| TCGA novel | 940 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091949051 | 940 | A>V | No | gnomAD | |
| rs2091949106 | 942 | G>V | No | Ensembl | |
| rs1557062523 | 948 | R>* | No | gnomAD | |
| rs372413645 | 948 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1557062526 | 950 | Q>R | No | gnomAD | |
| rs782094625 | 954 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs782693353 | 955 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs782693353 | 955 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs144666678 | 957 | N>H | No |
ESP TOPMed gnomAD |
|
| rs2091949329 | 958 | T>A | No | Ensembl | |
| rs782526023 | 958 | T>I | No |
ExAC gnomAD |
|
| rs2148424836 | 959 | S>I | No | Ensembl | |
| rs370374582 | 962 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091949427 | 962 | V>M | No | gnomAD | |
| TCGA novel | 963 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782543452 | 964 | G>D | No |
1000Genomes ExAC gnomAD |
|
| rs782233182 | 964 | G>S | No |
ExAC gnomAD |
|
| rs1191641733 | 966 | P>L | No | gnomAD | |
| rs782590898 | 969 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs782590898 | 969 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs781854042 | 971 | E>D | No |
ExAC gnomAD |
|
| rs782776023 | 971 | E>K | No |
ExAC gnomAD |
|
| rs782776023 | 971 | E>Q | No |
ExAC gnomAD |
|
| rs1416477659 | 973 | V>A | No |
TOPMed gnomAD |
|
| rs1185723830 | 975 | P>T | No |
TOPMed gnomAD |
|
| rs2091953489 | 976 | E>G | No | TOPMed | |
| rs375226965 | 976 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs375226965 | 976 | E>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782486603 | 977 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs139989610 | 978 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs369935750 | 979 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091953562 | 980 | C>S | No | TOPMed | |
|
COSM3733407 COSM3733409 COSM3733408 rs782508777 |
981 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
|
rs782400714 COSM1190647 |
981 | R>H | lung [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs782329303 | 983 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs782329303 | 983 | R>T | No |
ExAC TOPMed gnomAD |
|
| rs2091953676 | 986 | A>T | No | Ensembl | |
| rs2016507065 | 987 | A>T | No | TOPMed | |
| rs1557062662 | 987 | A>V | No | gnomAD | |
| rs782225065 | 989 | G>A | No |
ExAC gnomAD |
|
| rs782334605 | 992 | A>T | No |
ExAC gnomAD |
|
| rs781999056 | 992 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 994 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782060904 | 998 | G>D | No |
ExAC gnomAD |
|
| rs201369325 | 998 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2091953871 | 1001 | Q>H | No |
TOPMed gnomAD |
|
| rs782755267 | 1002 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs868912528 RCV000709843 |
1002 | R>H | No |
ClinVar Ensembl dbSNP |
|
| rs1557062686 | 1003 | T>I | No | gnomAD | |
| rs781826620 | 1005 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1461581650 | 1005 | L>P | No |
TOPMed gnomAD |
|
| rs189684685 | 1006 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1007 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091954049 | 1008 | P>S | No | TOPMed | |
|
COSM1466603 rs781907647 COSM5090368 COSM5090367 |
1010 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs373016226 | 1010 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091954113 | 1011 | Y>C | No | Ensembl | |
| rs376757604 | 1013 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1557062701 | 1017 | L>I | No | gnomAD | |
| rs201440646 | 1019 | A>E | No |
1000Genomes ExAC gnomAD |
|
| rs201440646 | 1019 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs782052955 | 1020 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs782272752 | 1023 | S>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1024 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782328918 | 1027 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM3747370 COSM3747371 rs782318928 |
1027 | R>W | liver [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs2148425608 | 1028 | G>R | No | Ensembl | |
| rs2091955916 | 1029 | G>D | No | TOPMed | |
| COSM1315328 | 1031 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
TCGA novel rs2091955939 |
1032 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs781971671 | 1033 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs782082528 | 1034 | R>C | No |
ExAC TOPMed gnomAD |
|
|
COSM4716004 COSM4716005 COSM4716003 rs181071909 |
1034 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs181071909 | 1034 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1557062766 | 1037 | G>D | No | gnomAD | |
| rs2148425863 | 1038 | T>A | No | Ensembl | |
| rs2091956054 | 1038 | T>I | No | Ensembl | |
| rs782803290 | 1039 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs781800371 | 1042 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs781800371 | 1042 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs782441783 | 1043 | V>M | No |
ExAC gnomAD |
|
| rs2091956210 | 1044 | Q>E | No | gnomAD | |
|
COSM5065937 COSM1569625 COSM5065936 rs781882423 |
1045 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs782090833 | 1045 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1433893121 | 1046 | P>S | No |
TOPMed gnomAD |
|
| rs2091956298 | 1050 | V>A | No | TOPMed | |
| rs1421049314 | 1053 | E>K | No | Ensembl | |
| rs371199032 | 1054 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs782450733 | 1056 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs782580336 | 1058 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs782580336 | 1058 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs782224778 | 1059 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs782341195 | 1060 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs782738305 | 1063 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1278546341 | 1064 | Q>P | No | TOPMed | |
| rs782245749 | 1065 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs782245749 | 1065 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs782245749 | 1065 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs2148425949 | 1066 | Q>K | No | Ensembl | |
| rs1557062831 | 1067 | D>N | No |
TOPMed gnomAD |
|
| rs1557062831 | 1067 | D>Y | No |
TOPMed gnomAD |
|
| rs2091956764 | 1071 | R>T | No | TOPMed | |
| rs782015410 | 1072 | R>K | No |
ExAC gnomAD |
|
| rs2148425969 | 1073 | S>T | No | Ensembl | |
| rs1293866821 | 1074 | C>Y | No | TOPMed | |
| rs782061585 | 1076 | A>T | No |
ExAC gnomAD |
|
| rs781964742 | 1079 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs782298822 | 1079 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs781964742 | 1079 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs782771749 | 1080 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1603247587 | 1081 | P>H | No |
TOPMed gnomAD |
|
| rs1603247587 | 1081 | P>R | No |
TOPMed gnomAD |
|
| rs781910392 | 1082 | Q>E | No |
ExAC gnomAD |
|
| rs2091957020 | 1083 | A>G | No | TOPMed | |
| rs1557062850 | 1083 | A>T | No | gnomAD | |
| rs186100707 | 1088 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782717211 | 1089 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs1189270790 | 1090 | G>R | No | TOPMed | |
| TCGA novel | 1091 | L>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557062860 | 1091 | L>M | No | gnomAD | |
| rs2091957196 | 1093 | Q>H | No | TOPMed | |
| rs1184422728 | 1094 | C>R | No | TOPMed | |
| rs1469582393 | 1095 | S>C | No | Ensembl | |
| rs1253237625 | 1096 | T>S | No |
TOPMed gnomAD |
|
| rs782125995 | 1097 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs374773455 | 1097 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs782775587 | 1100 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs781897955 | 1102 | S>L | No |
ExAC gnomAD |
|
| rs2091959274 | 1102 | S>P | No | TOPMed | |
|
rs111942299 RCV000887374 |
1104 | S>N | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1557062929 | 1104 | S>R | No | gnomAD | |
| rs111942299 | 1104 | S>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145360535 | 1109 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs145360535 | 1109 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs781786655 | 1109 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs958483288 | 1112 | A>T | No | Ensembl | |
| rs782552049 | 1113 | V>A | No |
ExAC gnomAD |
|
| rs1372997163 | 1113 | V>I | No | TOPMed | |
| rs2148426372 | 1114 | P>R | No | Ensembl | |
| rs782586367 | 1115 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2091959554 | 1115 | D>N | No | TOPMed | |
| rs782313304 | 1116 | R>T | No |
ExAC gnomAD |
|
| rs1455833743 | 1117 | A>G | No | gnomAD | |
| rs781969979 | 1117 | A>T | No |
ExAC gnomAD |
|
| rs782214419 | 1118 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs782391207 | 1119 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs782391207 | 1119 | P>R | No |
ExAC TOPMed gnomAD |
|
|
rs2148426396 TCGA novel |
1119 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1251959687 | 1121 | R>G | No |
TOPMed gnomAD |
|
| rs1196319451 | 1121 | R>Q | No |
TOPMed gnomAD |
|
|
rs1251959687 COSM1221303 COSM5064515 COSM5064514 |
1121 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs372502098 | 1122 | V>F | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1123 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM6186124 COSM6186123 COSM6186122 |
1124 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1603247794 | 1125 | T>A | No | Ensembl | |
| TCGA novel | 1126 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781934893 | 1129 | V>M | No |
ExAC TOPMed gnomAD |
|
|
COSM4844055 COSM4844056 COSM4844057 |
1131 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2148426437 | 1134 | A>G | No | Ensembl | |
| rs920485568 | 1134 | A>T | No |
TOPMed gnomAD |
|
| rs2054640225 | 1135 | S>R | No | TOPMed | |
| rs2148426443 | 1137 | S>N | No | Ensembl | |
| rs1557062965 | 1137 | S>R | No | gnomAD | |
| rs2091959959 | 1138 | G>E | No | Ensembl | |
| rs1603247804 | 1138 | G>R | No | TOPMed | |
| rs1348296727 | 1139 | G>D | No |
TOPMed gnomAD |
|
| rs2091959992 | 1140 | Q>K | No | Ensembl | |
| rs1557062967 | 1140 | Q>R | No | gnomAD | |
| rs1304470777 | 1141 | G>S | No |
TOPMed gnomAD |
|
|
COSM3973432 COSM3973431 COSM3973430 |
1145 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs375474840 | 1147 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs781818115 | 1147 | N>T | No |
ExAC gnomAD |
|
| rs782820984 | 1149 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs781889794 | 1149 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1396466475 | 1151 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1557062988 | 1152 | P>A | No | gnomAD | |
| rs1557062988 | 1152 | P>T | No | gnomAD | |
| rs782469872 | 1155 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs782469872 | 1155 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs782652825 | 1155 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs782652825 | 1155 | R>L | No |
ExAC TOPMed gnomAD |
|
|
rs6643791 VAR_061538 |
1156 | E>D | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs199905333 | 1156 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs892655372 | 1157 | G>A | No | Ensembl | |
| rs1417968010 | 1157 | G>R | No |
TOPMed gnomAD |
|
| rs781962579 | 1158 | P>L | No |
ExAC gnomAD |
|
| rs2091960448 | 1159 | A>T | No | Ensembl | |
| rs782348716 | 1160 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs146891755 | 1160 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM6118140 COSM6118142 COSM6118141 |
1160 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1557063009 | 1161 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1485879456 | 1163 | R>C | No |
TOPMed gnomAD |
|
| rs781979640 | 1163 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1557063020 | 1164 | L>F | No | gnomAD | |
| rs1557063022 | 1167 | G>S | No | gnomAD | |
| rs782712006 | 1169 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs782712006 | 1169 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs2091960668 COSM3559537 COSM3559538 COSM3559539 |
1171 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| TCGA novel | 1175 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557063091 | 1175 | E>K | No |
TOPMed gnomAD |
|
| rs1557063098 | 1176 | G>D | No |
TOPMed gnomAD |
|
|
COSM1659451 rs1356728532 COSM1659452 |
1176 | G>S | kidney [Cosmic] | No |
cosmic curated TOPMed |
| rs2091962525 | 1180 | G>A | No | TOPMed | |
| rs1557063102 | 1183 | K>R | No | gnomAD | |
| rs2091962581 | 1184 | E>K | No | TOPMed | |
| rs373692086 | 1185 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs781865230 | 1186 | V>M | No |
ExAC gnomAD |
|
| rs1557063113 | 1187 | R>C | No | gnomAD | |
| rs61741723 | 1187 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091962731 | 1188 | V>A | No | Ensembl | |
| rs782672041 | 1188 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs782672041 COSM3747374 COSM3747373 |
1188 | V>M | liver [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs868963669 | 1189 | H>N | No | 1000Genomes | |
| rs868963669 | 1189 | H>Y | No | 1000Genomes | |
| rs1381246310 | 1190 | I>V | No | Ensembl | |
| rs1557063131 | 1191 | G>S | No |
TOPMed gnomAD |
|
| rs201116168 | 1192 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs782623122 | 1192 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs377669094 | 1193 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs782162616 | 1194 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs781988597 | 1194 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1557063139 | 1195 | C>R | No |
TOPMed gnomAD |
|
| rs781932434 | 1199 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2091963051 | 1200 | L>F | No | TOPMed | |
| rs782741142 | 1201 | T>K | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1201 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs370950098 COSM4107831 COSM4107830 COSM4107832 |
1202 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs376345505 | 1202 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs376345505 | 1202 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs370950098 | 1202 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1335523852 | 1205 | L>V | No |
TOPMed gnomAD |
|
| rs782452845 | 1207 | C>F | No |
ExAC gnomAD |
|
| rs782707297 | 1208 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs782707297 | 1208 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs892512676 | 1209 | P>A | No |
TOPMed gnomAD |
|
| rs782475713 | 1209 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs892512676 | 1209 | P>S | No |
TOPMed gnomAD |
|
| rs868946447 | 1211 | A>V | No |
TOPMed gnomAD |
|
| rs782540666 | 1212 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2091963375 | 1212 | H>R | No | gnomAD | |
| rs377690692 | 1213 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs782190401 | 1214 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs781965227 | 1215 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1167177022 | 1215 | Q>R | No |
TOPMed gnomAD |
|
| rs1603248097 | 1217 | A>T | No | Ensembl | |
| rs370437111 | 1221 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2148427012 | 1223 | P>T | No | Ensembl | |
| rs781980952 | 1226 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1027526034 | 1229 | M>K | No |
TOPMed gnomAD |
|
| rs1027526034 | 1229 | M>T | No |
TOPMed gnomAD |
|
| rs1557063234 | 1229 | M>V | No | gnomAD | |
| TCGA novel | 1230 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091964710 | 1230 | G>S | No | TOPMed | |
| rs2091964744 | 1232 | V>M | No | TOPMed | |
| rs782242697 | 1239 | V>M | No |
ExAC gnomAD |
|
| rs782414899 | 1240 | Q>E | No | ExAC | |
| rs1443740375 | 1240 | Q>H | No | TOPMed | |
| rs2091964865 | 1240 | Q>L | No | gnomAD | |
| rs1355086646 | 1241 | Y>F | No | TOPMed | |
| rs1427606303 | 1242 | E>* | No |
TOPMed gnomAD |
|
| rs1427606303 | 1242 | E>Q | No |
TOPMed gnomAD |
|
| rs2091964974 | 1243 | A>P | No |
TOPMed gnomAD |
|
| rs2091964974 | 1243 | A>T | No |
TOPMed gnomAD |
|
| rs782181575 | 1244 | E>K | No |
ExAC gnomAD |
|
| rs2091965017 | 1245 | P>L | No | TOPMed | |
| rs782360679 | 1245 | P>S | No |
ExAC gnomAD |
|
| rs782360679 | 1245 | P>T | No |
ExAC gnomAD |
|
| rs782134820 | 1246 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs782157150 | 1248 | S>F | No |
TOPMed gnomAD |
|
| rs782152760 | 1248 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs782157150 | 1248 | S>Y | No |
TOPMed gnomAD |
|
|
CA415124975 RCV000512712 rs1557063267 |
1249 | A>T | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs781787378 | 1252 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs868924750 | 1252 | V>M | No |
TOPMed gnomAD |
|
| rs2091965233 | 1253 | E>K | No | Ensembl | |
| rs782718397 | 1254 | A>D | No |
ExAC gnomAD |
|
| rs782718397 | 1254 | A>V | No |
ExAC gnomAD |
|
| rs1557063276 | 1255 | Q>* | No |
TOPMed gnomAD |
|
| rs1557063276 | 1255 | Q>K | No |
TOPMed gnomAD |
|
| rs1471186672 | 1257 | G>D | No | gnomAD | |
| rs944707794 | 1257 | G>S | No | Ensembl | |
| rs1557063287 | 1258 | V>A | No |
TOPMed gnomAD |
|
| rs140301774 | 1258 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs782679434 | 1259 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs2091965428 | 1259 | G>S | No | Ensembl | |
| rs1482279481 | 1260 | M>T | No | TOPMed | |
| rs2091965495 | 1261 | G>D | No | gnomAD | |
| rs1282838584 | 1261 | G>S | No |
TOPMed gnomAD |
|
| rs781886576 | 1262 | A>T | No |
ExAC gnomAD |
|
| rs2091965546 | 1264 | V>A | No | Ensembl | |
| rs2091965602 | 1266 | I>M | No | TOPMed | |
| rs868919382 | 1267 | A>V | No | Ensembl | |
| rs150314931 | 1268 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1225021505 | 1271 | L>P | No |
TOPMed gnomAD |
|
| rs2091965769 | 1274 | L>F | No | Ensembl | |
| rs371794104 | 1275 | M>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs371794104 | 1275 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs781976995 | 1278 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2091968038 | 1278 | H>R | No | TOPMed | |
| rs1603248412 | 1279 | K>N | No | Ensembl | |
| rs1557063381 | 1281 | K>R | No | gnomAD | |
| rs1557063382 | 1282 | Q>E | No | gnomAD | |
| rs782284173 | 1282 | Q>R | No |
ExAC gnomAD |
|
| rs1557063389 | 1283 | A>G | No |
TOPMed gnomAD |
|
| rs1557063384 | 1283 | A>T | No | gnomAD | |
| rs1557063389 | 1283 | A>V | No |
TOPMed gnomAD |
|
| rs782396757 | 1285 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM4807696 COSM4807694 COSM4807695 |
1285 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2091968240 | 1287 | Y>C | No | TOPMed | |
| TCGA novel | 1288 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091968299 | 1290 | V>A | No | TOPMed | |
| rs1557063402 | 1292 | V>A | No | gnomAD | |
| rs1557063401 | 1292 | V>M | No | gnomAD | |
| rs966022575 | 1293 | Q>H | No | Ensembl | |
| rs1206334388 | 1298 | E>G | No | TOPMed | |
| rs1557063407 | 1299 | T>N | No | gnomAD | |
| rs1557063407 | 1299 | T>S | No | gnomAD | |
| rs2091968537 | 1300 | G>A | No |
TOPMed gnomAD |
|
| rs782819171 | 1300 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1319715193 | 1301 | V>M | No |
TOPMed gnomAD |
|
| rs1297524356 | 1302 | G>R | No | TOPMed | |
| rs2091968641 | 1304 | Q>H | No | TOPMed | |
| rs782694084 | 1305 | C>W | No |
ExAC TOPMed gnomAD |
|
| rs781835077 | 1306 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs201105227 | 1306 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2091968718 | 1307 | K>E | No | Ensembl | |
| rs1396741166 | 1307 | K>M | No | TOPMed | |
| rs1396741166 | 1307 | K>T | No | TOPMed | |
|
COSM1599024 COSM1117505 |
1311 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1557063454 | 1312 | L>F | No | gnomAD | |
| rs376295320 | 1313 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1373305496 | 1314 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1386280727 | 1316 | M>I | No | TOPMed | |
| rs1389267527 | 1317 | T>N | No |
TOPMed gnomAD |
|
| rs1557063465 | 1318 | D>H | No |
TOPMed gnomAD |
|
| rs1557063465 | 1318 | D>N | No |
TOPMed gnomAD |
|
| rs1557063470 | 1319 | L>H | No | gnomAD | |
| rs149518255 | 1321 | S>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1557063478 | 1321 | S>T | No | gnomAD | |
| rs782642884 | 1322 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs782642884 | 1322 | D>Y | No |
ExAC TOPMed gnomAD |
|
|
COSM1117506 COSM1599023 |
1323 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782405659 | 1324 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs374119831 | 1325 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs188069097 | 1325 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781827931 | 1327 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1027424282 | 1329 | P>R | No | TOPMed | |
| rs1482610323 | 1330 | F>L | No | Ensembl | |
| rs782036047 | 1332 | D>A | No |
ExAC gnomAD |
|
| rs782374659 | 1332 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs782142714 | 1334 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs782142714 | 1334 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs782709310 | 1334 | R>H | No |
ExAC gnomAD |
|
| rs138971273 | 1335 | T>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1232652447 | 1337 | A>T | No |
TOPMed gnomAD |
|
|
COSM4840780 rs200530402 COSM4840781 COSM137289 |
1338 | E>K | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP NCI-TCGA TOPMed gnomAD |
|
COSM4107833 rs781808141 COSM4107834 COSM4107835 |
1339 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs782437849 | 1339 | R>H | No |
ExAC TOPMed gnomAD |
|
|
COSM1648252 COSM755753 |
1340 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782206592 | 1340 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1557063526 | 1341 | F>I | No | Ensembl | |
| TCGA novel | 1343 | P>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1171592133 | 1344 | G>D | No |
TOPMed gnomAD |
|
| rs1557063533 | 1345 | H>Q | No | gnomAD | |
| rs782639579 | 1345 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs2148428061 | 1346 | G>S | No | Ensembl | |
| rs1557063538 | 1347 | G>A | No | gnomAD | |
| rs142851201 | 1347 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1557063541 | 1348 | C>Y | No | Ensembl | |
| rs966756251 | 1349 | P>L | No |
TOPMed gnomAD |
|
| rs966756251 | 1349 | P>R | No |
TOPMed gnomAD |
|
| rs1557063549 | 1354 | P>T | No | gnomAD | |
| rs1485864065 | 1355 | E>D | No |
TOPMed gnomAD |
|
| TCGA novel | 1355 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1355 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557063550 | 1356 | G>R | No | gnomAD | |
| TCGA novel | 1357 | P>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4399715 COSM4399716 rs1557063552 |
1359 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs781989622 | 1360 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1200018001 | 1360 | D>G | No |
TOPMed gnomAD |
|
| rs782004165 | 1361 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs782119260 | 1361 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs782004165 | 1361 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs781895505 | 1363 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs781895505 | 1363 | C>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1363 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1557063561 | 1365 | T>I | No | gnomAD | |
|
rs782068600 COSM3733318 COSM3733317 |
1367 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
rs782100931 COSM5134224 COSM5134225 |
1367 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs782068600 | 1367 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1341410243 | 1368 | Q>H | No |
TOPMed gnomAD |
|
|
COSM4107839 COSM4107840 |
1370 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201437211 | 1371 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201437211 | 1371 | T>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1660559683 | 1372 | Q>H | No | TOPMed | |
| rs2091971457 | 1373 | L>V | No | Ensembl | |
| rs1386669262 | 1375 | N>D | No | TOPMed | |
| rs2091971548 | 1377 | L>F | No | Ensembl | |
| rs1557063577 | 1378 | N>K | No | gnomAD | |
| rs1557063578 | 1379 | S>N | No | gnomAD | |
| rs1557063578 | 1379 | S>T | No | gnomAD | |
| rs782389181 | 1381 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199661205 | 1382 | F>L | No |
1000Genomes ExAC gnomAD |
|
| rs1557063585 | 1384 | L>F | No | gnomAD | |
| rs2148428187 | 1384 | L>P | No | Ensembl | |
| rs1557063587 | 1385 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1557063647 | 1387 | I>M | No | gnomAD | |
| rs2091973203 | 1387 | I>T | No | Ensembl | |
| rs2091973245 | 1390 | L>Q | No | Ensembl | |
| rs1412968429 | 1391 | E>G | No | TOPMed | |
| rs1557063653 | 1391 | E>K | No | gnomAD | |
| rs557107879 | 1392 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs557107879 | 1392 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1161282905 | 1394 | P>L | No |
TOPMed gnomAD |
|
|
RCV000879486 rs150648419 |
1395 | S>N | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs150648419 | 1395 | S>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2091973385 | 1397 | S>P | No | Ensembl | |
| rs966567963 | 1397 | S>Y | No | gnomAD | |
| rs1720499680 | 1398 | Q>R | No | Ensembl | |
| rs782548246 | 1399 | R>G | No |
ExAC gnomAD |
|
| rs782576689 | 1399 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs1000728365 | 1400 | D>N | No | Ensembl | |
| rs2091973523 | 1400 | D>V | No | Ensembl | |
| rs1189414694 | 1401 | R>C | No | TOPMed | |
|
rs782360488 COSM3764971 COSM3764970 |
1401 | R>H | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1557063661 | 1403 | H>Q | No |
TOPMed gnomAD |
|
| rs2091973576 | 1403 | H>R | No | TOPMed | |
| rs2091973614 | 1406 | S>L | No | gnomAD | |
| rs1557063665 | 1409 | S>L | No | gnomAD | |
| rs1603248832 | 1410 | L>P | No | Ensembl | |
| rs34054116 | 1411 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs34054116 | 1411 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782817411 | 1413 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs781918562 | 1414 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs781918562 | 1414 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs782718143 | 1417 | E>A | No | ExAC | |
| rs1557063681 | 1417 | E>Q | No | gnomAD | |
| rs782718143 | 1417 | E>V | No | ExAC | |
| rs2091973836 | 1418 | Y>H | No | TOPMed | |
| rs1305076624 | 1419 | L>M | No | TOPMed | |
| rs1436531616 | 1420 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1557063692 | 1421 | D>N | No | gnomAD | |
| rs782797156 | 1422 | I>V | No |
ExAC TOPMed |
|
| rs970937749 | 1423 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1321448343 | 1423 | M>T | No | TOPMed | |
| rs1557063700 | 1423 | M>V | No | gnomAD | |
| rs781811393 | 1424 | R>S | No |
ExAC gnomAD |
|
|
COSM1599021 COSM1117508 |
1425 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781826425 | 1428 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs2091974138 | 1429 | D>E | No | Ensembl | |
| rs781899470 | 1430 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs370364112 | 1431 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1048660836 | 1432 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs2091974284 | 1433 | H>R | No | Ensembl | |
| rs1388549943 | 1433 | H>Y | No | TOPMed | |
| rs1557063713 | 1434 | Y>D | No | gnomAD | |
| TCGA novel | 1434 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1221304 rs782577261 |
1435 | V>M | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1557063719 | 1437 | R>K | No | gnomAD | |
| TCGA novel | 1440 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782177378 | 1440 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs2091974452 | 1441 | L>F | No | Ensembl | |
| rs1557063722 | 1442 | M>L | No | gnomAD | |
| rs2091974503 | 1442 | M>T | No | Ensembl | |
| rs1557063722 | 1442 | M>V | No | gnomAD | |
| rs2091974515 | 1444 | R>C | No | gnomAD | |
| rs782352445 | 1444 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs2091974555 | 1445 | R>K | No | Ensembl | |
| rs2148429000 | 1446 | T>K | No | Ensembl | |
|
TCGA novel rs782278086 |
1447 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC TOPMed gnomAD |
| rs782331949 | 1449 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1159021442 | 1449 | M>V | No |
TOPMed gnomAD |
|
| rs1557063802 | 1451 | E>* | No | gnomAD | |
| rs368710818 | 1451 | E>D | No |
ESP TOPMed |
|
| rs2091977353 | 1451 | E>G | No | TOPMed | |
| rs781921372 | 1455 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1557063807 | 1455 | T>S | No | gnomAD | |
| rs1557063809 | 1459 | S>P | No | gnomAD | |
| rs2091977432 | 1461 | C>S | No | TOPMed | |
| rs2091977447 | 1462 | L>Q | No | TOPMed | |
| rs782006902 | 1464 | A>G | No |
ExAC TOPMed gnomAD |
|
|
COSM1221305 COSM5164448 rs782351638 |
1464 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs782115691 | 1467 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs144474723 | 1468 | E>K | No | ESP | |
| rs2091980710 | 1468 | E>V | No | TOPMed | |
| rs2091980766 | 1469 | V>A | No | TOPMed | |
| rs375875993 | 1469 | V>L | No |
ESP TOPMed gnomAD |
|
| rs375875993 | 1469 | V>M | No |
ESP TOPMed gnomAD |
|
| rs1557063906 | 1470 | A>G | No | gnomAD | |
| rs1310280930 | 1470 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1557063906 | 1470 | A>V | No | gnomAD | |
| TCGA novel | 1471 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091980812 | 1473 | P>S | No | Ensembl | |
| rs146671037 | 1474 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs782730938 | 1476 | M>L | No |
ExAC gnomAD |
|
| rs782730938 | 1476 | M>V | No |
ExAC gnomAD |
|
| rs781997851 | 1478 | F>L | No |
ExAC gnomAD |
|
| rs1369583250 | 1479 | R>W | No | TOPMed | |
| TCGA novel | 1480 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091980982 | 1481 | I>T | No | gnomAD | |
| rs1557063914 | 1482 | Q>H | No |
TOPMed gnomAD |
|
| rs1467132797 | 1482 | Q>R | No | TOPMed | |
| rs2091981051 | 1483 | Y>H | No | TOPMed | |
| rs782817526 | 1484 | Q>E | No |
ExAC gnomAD |
|
| rs781810671 | 1486 | D>E | No | ExAC | |
| rs782453757 | 1488 | G>D | No |
ExAC gnomAD |
|
| rs2091981121 | 1489 | P>L | No | gnomAD | |
| rs2091981170 | 1490 | V>A | No | TOPMed | |
| rs1557063925 | 1490 | V>M | No | gnomAD | |
| rs2091981187 | 1492 | A>T | No |
TOPMed gnomAD |
|
| rs2091981219 | 1493 | V>M | No |
TOPMed gnomAD |
|
|
COSM1599020 COSM1117510 |
1495 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781830053 | 1495 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs782640225 | 1497 | A>S | No |
ExAC gnomAD |
|
| rs782640225 | 1497 | A>T | No |
ExAC gnomAD |
|
| rs868939578 | 1499 | R>Q | No | TOPMed | |
| rs2091981365 | 1503 | D>N | No | TOPMed | |
| rs1482580213 | 1505 | R>C | No |
TOPMed gnomAD |
|
|
COSM1221302 rs782590076 |
1505 | R>H | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1194232949 | 1508 | R>Q | No |
TOPMed gnomAD |
|
|
rs2091981458 COSM1599019 COSM1117511 |
1508 | R>W | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs984250524 | 1509 | E>K | No |
TOPMed gnomAD |
|
| rs1018547890 | 1510 | D>H | No |
TOPMed gnomAD |
|
| rs1018547890 | 1510 | D>N | No |
TOPMed gnomAD |
|
| rs782539109 | 1511 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782539109 | 1511 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1557063960 | 1512 | E>Q | No | gnomAD | |
| rs964681482 | 1513 | F>I | No | Ensembl | |
| rs1557063969 | 1515 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
|
rs782071212 COSM457058 |
1517 | T>M | breast [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1290700778 | 1519 | M>T | No |
TOPMed gnomAD |
|
| rs781970640 | 1521 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs782081945 | 1523 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs782172275 | 1525 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs781861924 | 1525 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs782172275 | 1525 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1557064003 | 1526 | A>T | No |
1000Genomes TOPMed gnomAD |
|
| rs2091981985 | 1527 | G>V | No | Ensembl | |
| rs1557064013 | 1528 | G>E | No | gnomAD | |
| rs1557064008 | 1528 | G>R | No |
TOPMed gnomAD |
|
| rs1603249350 | 1529 | A>V | No | TOPMed | |
| rs1402462072 | 1530 | A>P | No | Ensembl | |
| rs1402462072 | 1530 | A>T | No | Ensembl | |
|
COSM1117512 COSM1599018 |
1531 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782602150 | 1532 | S>N | No | ExAC | |
| rs879986670 | 1534 | E>K | No | gnomAD | |
| rs782641437 | 1535 | M>I | No |
ExAC TOPMed gnomAD |
|
|
VAR_019682 rs5987155 |
1535 | M>T | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1414126396 | 1536 | Q>L | No |
TOPMed gnomAD |
|
| rs1414126396 | 1536 | Q>R | No |
TOPMed gnomAD |
|
| rs369982782 | 1537 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs372579744 | 1537 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs372579744 | 1537 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs369982782 | 1537 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1189346572 | 1538 | V>M | No |
TOPMed gnomAD |
|
| rs1557064050 | 1540 | A>V | No | gnomAD | |
| rs1257094238 | 1541 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs148020794 | 1541 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1557064062 | 1542 | V>L | No |
TOPMed gnomAD |
|
| rs1557064063 | 1543 | L>F | No | gnomAD | |
| rs782000261 | 1544 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1603249387 | 1545 | T>M | No | Ensembl | |
| rs782420793 | 1548 | I>V | No |
ExAC gnomAD |
|
| rs1557064077 | 1549 | T>I | No |
TOPMed gnomAD |
|
| rs1557064077 | 1549 | T>N | No |
TOPMed gnomAD |
|
| rs1270580199 | 1553 | E>K | No |
TOPMed gnomAD |
|
| rs1209839776 | 1554 | K>E | No | TOPMed | |
| rs1329059623 | 1554 | K>M | No | TOPMed | |
| rs1557064090 | 1555 | V>E | No | gnomAD | |
| rs1569542156 | 1556 | L>S | No | Ensembl | |
|
COSM3559540 COSM3559541 |
1558 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782695147 | 1559 | V>D | No |
ExAC TOPMed gnomAD |
|
| rs375753845 | 1559 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1557064101 | 1560 | Y>* | No |
TOPMed gnomAD |
|
| rs868906899 | 1562 | G>D | No | Ensembl | |
| rs368893732 | 1562 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs782073828 | 1563 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1603249425 | 1563 | T>P | No | Ensembl | |
| TCGA novel | 1563 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1603249432 | 1565 | F>L | No | Ensembl | |
| rs1311512031 | 1567 | Q>E | No | TOPMed | |
| rs2091983168 | 1567 | Q>H | No | TOPMed | |
| rs1449877215 | 1568 | R>K | No |
TOPMed gnomAD |
|
| rs2091983189 | 1568 | R>W | No | TOPMed | |
| rs1390117971 | 1572 | H>N | No | TOPMed | |
| rs1321157013 | 1573 | A>S | No |
TOPMed gnomAD |
|
| rs1321157013 | 1573 | A>T | No |
TOPMed gnomAD |
|
| rs781853080 | 1573 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs782413172 | 1578 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA gnomAD |
|
rs1557064222 COSM5130782 COSM1466607 |
1579 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
|
rs1343910080 COSM1683005 COSM1683006 |
1579 | R>H | large_intestine [Cosmic] | No |
cosmic curated TOPMed |
| rs1343910080 | 1579 | R>L | No | TOPMed | |
| rs782484213 | 1583 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs201042784 | 1584 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 1585 | H>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs868937028 | 1586 | L>M | No |
TOPMed gnomAD |
|
| rs782257618 | 1587 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1557064241 | 1589 | S>* | No | gnomAD | |
| rs1557064241 | 1589 | S>L | No | gnomAD | |
| rs1557064241 | 1589 | S>W | No | gnomAD | |
| rs145978031 | 1591 | E>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2091986563 | 1592 | D>N | No | Ensembl | |
| rs1603249646 | 1594 | T>P | No | Ensembl | |
| rs146832392 | 1596 | V>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146832392 | 1596 | V>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs138779643 | 1596 | V>L | No |
ESP ExAC gnomAD |
|
| rs138779643 | 1596 | V>M | No |
ESP ExAC gnomAD |
|
| rs782173689 | 1598 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1603249659 | 1599 | N>T | No | Ensembl | |
| rs782803873 | 1599 | N>Y | No |
ExAC TOPMed gnomAD |
|
| rs782437322 | 1600 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs781808946 | 1600 | H>Y | No |
ExAC gnomAD |
|
| rs782755006 | 1601 | W>* | No |
ExAC TOPMed gnomAD |
|
| rs200030126 | 1602 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs782627655 | 1603 | R>I | No |
ExAC TOPMed gnomAD |
|
| rs782627655 | 1603 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs1603249670 | 1603 | R>S | No | Ensembl | |
| rs782295733 | 1604 | L>F | No |
ExAC gnomAD |
|
| rs782295733 | 1604 | L>V | No |
ExAC gnomAD |
|
|
rs147439766 RCV000996045 |
1606 | T>I | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1603249681 | 1606 | T>P | No | Ensembl | |
| rs199599060 | 1607 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1603249690 | 1608 | Q>* | No | Ensembl | |
| rs782244373 | 1608 | Q>H | No |
ExAC gnomAD |
|
| rs2091987154 | 1609 | H>Y | No | Ensembl | |
| rs1557064265 | 1610 | Y>* | No | gnomAD | |
| rs1603249697 | 1611 | K>R | No | Ensembl | |
| rs1557064574 | 1613 | P>A | No | gnomAD | |
| rs2092003216 | 1616 | A>E | No | TOPMed | |
| rs2092003216 | 1616 | A>G | No | TOPMed | |
| rs1557064575 | 1617 | T>I | No | gnomAD | |
|
COSM4107844 COSM4107843 |
1618 | V>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1557064576 | 1618 | V>M | No | gnomAD | |
|
rs1557064580 COSM3372151 COSM3372152 |
1620 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs782241170 | 1621 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1402760182 | 1623 | Q>R | No | TOPMed | |
| rs1557064587 | 1624 | L>P | No |
TOPMed gnomAD |
|
| rs1557064593 | 1625 | H>R | No |
TOPMed gnomAD |
|
| rs1408267129 | 1625 | H>Y | No |
TOPMed gnomAD |
|
| rs782507457 | 1626 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA TOPMed gnomAD |
| rs782668477 | 1626 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1244516232 | 1628 | S>N | No | TOPMed | |
| rs2148432717 | 1630 | I>F | No | Ensembl | |
| rs1460853789 | 1633 | S>N | No |
TOPMed gnomAD |
|
| rs2092003673 | 1634 | L>P | No | Ensembl | |
| rs2092003649 | 1634 | L>V | No | Ensembl | |
| rs782271975 | 1635 | A>V | No |
ExAC gnomAD |
|
| TCGA novel | 1636 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782318199 | 1636 | Q>K | No |
ExAC gnomAD |
|
| rs1557064612 | 1641 | G>R | No | gnomAD | |
| rs191126455 | 1642 | E>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1317223148 | 1642 | E>K | No |
TOPMed gnomAD |
|
| rs1223493587 | 1643 | N>Y | No | TOPMed | |
| rs782424228 | 1645 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs782023871 | 1646 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2092007481 | 1647 | L>M | No | TOPMed | |
| rs1284313486 | 1650 | G>S | No |
TOPMed gnomAD |
|
|
VAR_068807 rs34762690 |
1651 | E>A | No |
UniProt Ensembl dbSNP |
|
| rs34762690 | 1651 | E>G | No | Ensembl | |
| rs201844222 | 1651 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs782087499 | 1652 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1569542197 | 1652 | E>G | No | TOPMed | |
| rs781974282 | 1653 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs781974282 | 1653 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs782787926 | 1653 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs781974282 | 1653 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs782787926 | 1653 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs782790314 | 1654 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs782790314 | 1654 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1557064736 | 1655 | V>A | No | gnomAD | |
|
COSM4612518 COSM1466608 rs782076287 |
1655 | V>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs782445875 | 1655 | V>M | No |
ExAC gnomAD |
|
| rs2148433393 | 1659 | H>Q | No | Ensembl | |
| rs1557064744 | 1661 | V>A | No | gnomAD | |
| rs1603250800 | 1663 | A>T | No | Ensembl | |
| rs1265472852 | 1664 | T>N | No |
TOPMed gnomAD |
|
| rs1265472852 | 1664 | T>S | No |
TOPMed gnomAD |
|
|
rs372327696 COSM1466609 COSM4911192 |
1665 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA gnomAD |
| rs782682576 | 1666 | E>A | No |
ExAC gnomAD |
|
| rs1179057801 | 1667 | P>S | No |
TOPMed gnomAD |
|
| rs2092008124 | 1668 | E>A | No | Ensembl | |
| rs1480962965 | 1668 | E>K | No | TOPMed | |
| rs1251258499 | 1669 | G>R | No | TOPMed | |
| rs1251258499 | 1669 | G>W | No | TOPMed | |
| rs781957820 | 1670 | A>V | No |
TOPMed gnomAD |
|
| rs781924481 | 1671 | K>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200641103 | 1672 | V>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200641103 | 1672 | V>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782575528 | 1672 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs782348820 | 1673 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1391474144 | 1673 | R>W | No |
TOPMed gnomAD |
|
|
COSM3780390 COSM3780389 rs782412893 |
1678 | R>Q | pancreas [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
rs782236615 COSM5426767 COSM5426768 |
1678 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1557064787 | 1679 | E>D | No | gnomAD | |
| rs199536649 | 1680 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs199536649 | 1680 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782063016 | 1680 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs199536649 | 1680 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782308553 | 1681 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1557064796 | 1682 | P>S | No |
TOPMed gnomAD |
|
| rs2092008657 | 1684 | R>G | No | Ensembl | |
| rs2092008677 | 1684 | R>M | No | TOPMed | |
| rs369074199 | 1686 | K>N | No |
ESP TOPMed gnomAD |
|
| rs1161484014 | 1687 | A>G | No |
TOPMed gnomAD |
|
| rs782079909 | 1687 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs782079909 | 1687 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs1557064802 | 1689 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs781902036 | 1692 | Y>C | No |
ExAC gnomAD |
|
| rs1249993822 | 1694 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1557064811 | 1695 | R>H | No | gnomAD | |
| rs1557064816 | 1698 | S>P | No | gnomAD | |
| rs1557064818 | 1699 | M>V | No | gnomAD | |
| rs782055888 | 1702 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2092010780 | 1704 | Q>K | No | TOPMed | |
| rs372812213 | 1705 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM5153794 COSM5153793 COSM1466610 |
1705 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs570470027 | 1706 | F>L | No | Ensembl | |
| rs1557064882 | 1707 | V>A | No | gnomAD | |
| rs781894023 | 1708 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1557064884 | 1709 | D>N | No | gnomAD | |
| rs144227320 | 1710 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs148745452 | 1712 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs2092011076 | 1713 | A>T | No | Ensembl | |
| rs1557064890 | 1714 | I>V | No | gnomAD | |
| rs2092011116 | 1716 | S>G | No | TOPMed | |
| rs782483104 | 1716 | S>N | No |
ExAC gnomAD |
|
| rs782483104 | 1716 | S>T | No |
ExAC gnomAD |
|
| rs142300328 | 1717 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs142300328 COSM1466611 |
1717 | V>M | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs782202818 | 1718 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1426766177 | 1719 | R>Q | No |
TOPMed gnomAD |
|
| rs782674766 | 1719 | R>W | No |
ExAC gnomAD |
|
| rs2092011339 | 1722 | P>S | No | TOPMed | |
| rs1366341464 | 1723 | I>V | No | TOPMed | |
|
COSM4107848 COSM4107847 |
1724 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs377395548 | 1724 | A>T | No |
ESP TOPMed gnomAD |
|
| rs2092011428 | 1724 | A>V | No | Ensembl | |
| rs1557064919 | 1725 | V>A | No | gnomAD | |
| rs781971067 | 1725 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1375236498 | 1728 | L>V | No |
TOPMed gnomAD |
|
| rs1557064929 | 1729 | F>S | No | gnomAD | |
|
COSM1490649 COSM5231017 |
1730 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1194383256 | 1730 | D>N | No | TOPMed | |
| rs2092011617 | 1731 | L>F | No | Ensembl | |
| rs782175065 | 1733 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs2092011722 | 1734 | E>D | No | TOPMed | |
| rs1557064940 | 1734 | E>K | No |
TOPMed gnomAD |
|
| rs1557064942 | 1735 | L>P | No | gnomAD | |
| rs782778557 | 1737 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1224327874 | 1737 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs782041663 | 1739 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1341418700 | 1740 | G>D | No | TOPMed | |
| rs2092011916 | 1740 | G>S | No | TOPMed | |
| rs370954582 | 1742 | E>K | No |
ESP TOPMed gnomAD |
|
| rs370954582 | 1742 | E>Q | No |
ESP TOPMed gnomAD |
|
| rs782757038 | 1745 | G>R | No |
ExAC gnomAD |
|
| rs934807409 | 1746 | T>I | No | Ensembl | |
| rs1557064966 | 1749 | I>M | No |
TOPMed gnomAD |
|
| rs1284950509 | 1752 | T>S | No | TOPMed | |
|
COSM3236026 COSM3236025 rs1557065078 |
1758 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM4762847 COSM4762846 rs1557065075 |
1758 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1557065083 | 1760 | W>* | No | gnomAD | |
| rs201223941 | 1761 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs914741321 | 1763 | A>S | No | gnomAD | |
| rs914741321 | 1763 | A>T | No | gnomAD | |
| TCGA novel | 1764 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782519182 | 1765 | K>N | No |
ExAC TOPMed gnomAD |
|
|
COSM1117516 COSM1599014 rs782688715 |
1766 | N>S | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1408194532 | 1767 | P>A | No | TOPMed | |
| rs782563364 | 1769 | L>F | No |
ExAC gnomAD |
|
| rs373926896 | 1770 | I>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs782334408 | 1772 | D>A | No |
ExAC gnomAD |
|
|
COSM1599013 COSM1117517 |
1773 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs139661833 | 1774 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs368444846 | 1775 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs368444846 | 1775 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1184187177 | 1776 | S>L | No |
TOPMed gnomAD |
|
| rs2092015726 | 1778 | N>H | No | Ensembl | |
| rs781953533 | 1778 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs2092015744 | 1778 | N>S | No | TOPMed | |
| rs782133606 | 1779 | V>M | No |
ExAC gnomAD |
|
| TCGA novel | 1781 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs371914178 | 1781 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1217660395 | 1783 | L>F | No | TOPMed | |
| rs781982608 | 1783 | L>H | No | Ensembl | |
| rs1448968248 | 1784 | A>V | No |
TOPMed gnomAD |
|
| rs1557065129 | 1786 | I>V | No | Ensembl | |
| rs1003955955 | 1787 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| TCGA novel | 1788 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781909364 | 1789 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1286924099 | 1789 | T>P | No |
TOPMed gnomAD |
|
| rs375450207 | 1791 | I>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs375450207 | 1791 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs782152232 | 1791 | I>V | No | ExAC | |
| rs2092016100 | 1796 | T>I | No |
TOPMed gnomAD |
|
|
COSM1117518 rs781784671 COSM1599012 |
1797 | S>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs202202015 | 1798 | E>K | No |
TOPMed gnomAD |
|
| rs1379068546 | 1801 | V>M | No |
TOPMed gnomAD |
|
| rs781865529 | 1802 | G>D | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1803 | R>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2092016273 | 1803 | R>Q | No | TOPMed | |
| rs368531489 | 1803 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1557065203 | 1804 | D>N | No | gnomAD | |
| rs1557065207 | 1809 | K>E | No | gnomAD | |
|
COSM4107850 COSM4107849 COSM4107851 |
1809 | K>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM1648250 COSM755751 COSM1648249 |
1809 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782547073 | 1810 | L>R | No |
ExAC gnomAD |
|
| rs1303243413 | 1813 | A>T | No |
TOPMed gnomAD |
|
| rs369580403 | 1814 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1456905839 | 1814 | R>W | No | TOPMed | |
| rs2092018479 | 1815 | E>D | No |
TOPMed gnomAD |
|
| rs2092018500 | 1817 | P>T | No | Ensembl | |
|
COSM5178342 COSM1466612 rs782313955 |
1818 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1410439897 | 1818 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs782313955 | 1818 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1728515858 | 1826 | Y>D | No | TOPMed | |
| rs1557065300 | 1828 | A>S | No | gnomAD | |
| rs782253397 | 1828 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1557065303 | 1829 | D>G | No | gnomAD | |
| TCGA novel | 1829 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs146644526 | 1831 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146644526 | 1831 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782021262 | 1831 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs2092019981 | 1832 | Q>H | No | Ensembl | |
| rs374980397 | 1832 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1031655394 | 1833 | S>N | No | Ensembl | |
| rs1265441530 | 1834 | S>F | No | TOPMed | |
| rs782354124 | 1835 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs782354124 | 1835 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs139986679 | 1836 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1310065775 | 1841 | M>T | No |
TOPMed gnomAD |
|
| rs367877344 | 1842 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2148435306 | 1843 | S>Y | No | Ensembl | |
| rs1240162355 | 1847 | E>K | No |
TOPMed gnomAD |
|
| rs1329168360 | 1850 | G>R | No |
TOPMed gnomAD |
|
| rs782149290 | 1852 | Y>F | No |
ExAC gnomAD |
|
| rs1557065407 | 1853 | T>A | No | gnomAD | |
| rs782709924 | 1853 | T>I | No |
ExAC TOPMed gnomAD |
|
|
rs939639089 COSM1315330 |
1854 | S>C | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs1224000962 | 1856 | P>H | No | TOPMed | |
| rs1557065415 | 1856 | P>T | No | gnomAD | |
|
COSM4107853 COSM4107852 |
1858 | C>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2092022696 | 1858 | C>S | No | Ensembl | |
| rs781790936 | 1860 | E>K | No |
ExAC gnomAD |
|
| TCGA novel | 1864 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782617221 | 1869 | I>T | No |
TOPMed gnomAD |
|
| rs368280793 | 1870 | H>R | No |
ESP ExAC TOPMed |
|
| rs149060321 | 1870 | H>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs898204288 | 1871 | R>K | No | Ensembl | |
| rs1557065436 | 1872 | Y>C | No | gnomAD | |
| rs1690948629 | 1873 | Y>C | No | TOPMed | |
| rs1557065439 | 1873 | Y>H | No | gnomAD | |
| rs781961323 | 1876 | I>V | No | 1000Genomes | |
| rs1557065539 | 1877 | I>L | No | gnomAD | |
| rs145843639 | 1880 | L>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2092025538 | 1881 | E>K | No |
TOPMed gnomAD |
|
| rs1345961786 | 1882 | E>G | No |
TOPMed gnomAD |
|
| rs1603251867 | 1882 | E>K | No | Ensembl | |
| rs1274350639 | 1883 | D>E | No | TOPMed | |
| rs781831904 | 1884 | P>H | No |
ExAC gnomAD |
|
| rs782685997 | 1884 | P>S | No |
ExAC gnomAD |
|
| rs1231077936 | 1886 | G>D | No | TOPMed | |
| rs1379804084 | 1887 | Q>P | No |
TOPMed gnomAD |
|
| rs1557065554 | 1889 | L>P | No |
TOPMed gnomAD |
|
| rs2092025778 | 1890 | Q>R | No | Ensembl | |
| rs908101495 | 1891 | L>M | No |
TOPMed gnomAD |
|
| rs908101495 | 1891 | L>V | No |
TOPMed gnomAD |
|
| rs2092025838 | 1892 | A>V | No | Ensembl | |
| rs2148436065 | 1893 | C>S | No | Ensembl | |
| rs782240444 | 1894 | R>C | No |
ExAC TOPMed gnomAD |
|
|
COSM3694430 rs973767706 COSM3694429 |
1894 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs973767706 | 1894 | R>L | No | gnomAD | |
| rs1414736407 | 1896 | Q>R | No |
TOPMed gnomAD |
|
| rs371248419 | 1899 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs782194744 | 1900 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs782673164 | 1900 | A>T | No |
ExAC gnomAD |
|
| rs782194744 | 1900 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1557065587 | 1902 | V>L | No | gnomAD | |
| rs782313067 | 1903 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs782313067 | 1903 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs782323338 | 1904 | N>K | No |
1000Genomes TOPMed gnomAD |
|
| rs2092026228 | 1908 | D>N | No | TOPMed | |
| rs1177987495 | 1909 | L>P | No |
TOPMed gnomAD |
|
| rs1177987495 | 1909 | L>R | No |
TOPMed gnomAD |
No associated diseases with Q9ULL4
13 regional properties for Q9ULL4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Sema domain | 31 - 471 | IPR001627 |
| repeat | Plexin repeat | 474 - 525 | IPR002165 |
| domain | IPT domain | 834 - 925 | IPR002909-1 |
| domain | IPT domain | 926 - 1012 | IPR002909-2 |
| domain | IPT domain | 1014 - 1145 | IPR002909-3 |
| domain | IPT domain | 1167 - 1242 | IPR002909-4 |
| domain | Plexin, cytoplasmic RasGAP domain | 1327 - 1876 | IPR013548 |
| domain | PSI domain | 473 - 526 | IPR016201-1 |
| domain | PSI domain | 620 - 682 | IPR016201-2 |
| domain | PSI domain | 787 - 833 | IPR016201-3 |
| domain | Plexin, TIG domain 1 | 533 - 619 | IPR041019 |
| domain | Plexin, TIG domain 2 | 691 - 786 | IPR041362 |
| domain | Plexin, cytoplasmic RhoGTPase-binding domain | 1501 - 1618 | IPR046800 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell surface | The external part of the cell wall and/or plasma membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| semaphorin receptor complex | A stable binary complex of a neurophilin and a plexin, together forming a functional semaphorin receptor. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| cell-cell adhesion mediator activity | The binding by a cell-adhesion protein on the cell surface to an extracellular matrix component, to mediate adhesion of the cell to another cell. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| Rho GDP-dissociation inhibitor binding | Binding to a Rho GDP-dissociation inhibitor protein. |
| semaphorin receptor activity | Combining with a semaphorin, and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| cell chemotaxis | The directed movement of a motile cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| homophilic cell adhesion via plasma membrane adhesion molecules | The attachment of a plasma membrane adhesion molecule in one cell to an identical molecule in an adjacent cell. |
| negative regulation of cell adhesion | Any process that stops, prevents, or reduces the frequency, rate or extent of cell adhesion. |
| negative regulation of cell migration | Any process that stops, prevents, or reduces the frequency, rate or extent of cell migration. |
| negative regulation of GTPase activity | Any process that stops or reduces the rate of GTP hydrolysis by a GTPase. |
| negative regulation of lamellipodium assembly | Any process that decreases the rate, frequency or extent of the formation of a lamellipodium, a thin sheetlike extension of the surface of a migrating cell. |
| positive chemotaxis | The directed movement of a motile cell or organism towards a higher concentration of a chemical. |
| positive regulation of axonogenesis | Any process that activates or increases the frequency, rate or extent of axonogenesis. |
| positive regulation of endothelial cell proliferation | Any process that activates or increases the rate or extent of endothelial cell proliferation. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| semaphorin-plexin signaling pathway | The series of molecular signals generated as a consequence of a semaphorin receptor (composed of a plexin and a neurophilin) binding to a semaphorin ligand. |
| semaphorin-plexin signaling pathway involved in axon guidance | Any semaphorin-plexin signaling pathway that is involved in axon guidance. |
27 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q769I5 | MET | Hepatocyte growth factor receptor | Bos taurus (Bovine) | PR |
| A0M8S8 | MET | Hepatocyte growth factor receptor | Felis catus (Cat) (Felis silvestris catus) | PR |
| Q04912 | MST1R | Macrophage-stimulating protein receptor | Homo sapiens (Human) | EV |
| P08581 | MET | Hepatocyte growth factor receptor | Homo sapiens (Human) | EV |
| O75051 | PLXNA2 | Plexin-A2 | Homo sapiens (Human) | SS |
| Q9UIW2 | PLXNA1 | Plexin-A1 | Homo sapiens (Human) | EV SS |
| P51805 | PLXNA3 | Plexin-A3 | Homo sapiens (Human) | SS |
| O15031 | PLXNB2 | Plexin-B2 | Homo sapiens (Human) | SS |
| O43157 | PLXNB1 | Plexin-B1 | Homo sapiens (Human) | EV SS |
| Q9HCM2 | PLXNA4 | Plexin-A4 | Homo sapiens (Human) | SS |
| P70208 | Plxna3 | Plexin-A3 | Mus musculus (Mouse) | EV SS |
| Q3UH93 | Plxnd1 | Plexin-D1 | Mus musculus (Mouse) | SS |
| P70207 | Plxna2 | Plexin-A2 | Mus musculus (Mouse) | SS |
| Q62190 | Mst1r | Macrophage-stimulating protein receptor | Mus musculus (Mouse) | SS |
| B2RXS4 | Plxnb2 | Plexin-B2 | Mus musculus (Mouse) | SS |
| Q8CJH3 | Plxnb1 | Plexin-B1 | Mus musculus (Mouse) | SS |
| Q9QZC2 | Plxnc1 | Plexin-C1 | Mus musculus (Mouse) | SS |
| Q80UG2 | Plxna4 | Plexin-A4 | Mus musculus (Mouse) | SS |
| P16056 | Met | Hepatocyte growth factor receptor | Mus musculus (Mouse) | PR |
| P70206 | Plxna1 | Plexin-A1 | Mus musculus (Mouse) | EV SS |
| Q9QY40 | Plxnb3 | Plexin-B3 | Mus musculus (Mouse) | SS |
| Q2QLE0 | MET | Hepatocyte growth factor receptor | Sus scrofa (Pig) | PR |
| P97523 | Met | Hepatocyte growth factor receptor | Rattus norvegicus (Rat) | PR |
| D3ZPX4 | Plxna3 | Plexin-A3 | Rattus norvegicus (Rat) | SS |
| D3ZLH5 | Plxnb3 | Plexin-B3 | Rattus norvegicus (Rat) | SS |
| Q6BEA0 | plxna4 | Plexin-A4 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| B0S5N4 | plxna3 | Plexin A3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MCHAAQETPL | LHHFMAPVMA | RWPPFGLCLL | LLLLSPPPLP | LTGAHRFSAP | NTTLNHLALA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PGRGTLYVGA | VNRLFQLSPE | LQLEAVAVTG | PVIDSPDCVP | FRDPAECPQA | QLTDNANQLL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LVSSRAQELV | ACGQVRQGVC | ETRRLGDVAE | VLYQAEDPGD | GQFVAANTPG | VATVGLVVPL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PGRDLLLVAR | GLAGKLSAGV | PPLAIRQLAG | SQPFSSEGLG | RLVVGDFSDY | NNSYVGAFAD |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ARSAYFVFRR | RGARAQAEYR | SYVARVCLGD | TNLYSYVEVP | LACQGQGLIQ | AAFLAPGTLL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GVFAAGPRGT | QAALCAFPMV | ELGASMEQAR | RLCYTAGGRG | PSGAEEATVE | YGVTSRCVTL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PLDSPESYPC | GDEHTPSPIA | GRQPLEVQPL | LKLGQPVSAV | AALQADGHMI | AFLGDTQGQL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| YKVFLHGSQG | QVYHSQQVGP | PGSAISPDLL | LDSSGSHLYV | LTAHQVDRIP | VAACPQFPDC |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ASCLQAQDPL | CGWCVLQGRC | TRKGQCGRAG | QLNQWLWSYE | EDSHCLHIQS | LLPGHHPRQE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| QGQVTLSVPR | LPILDADEYF | HCAFGDYDSL | AHVEGPHVAC | VTPPQDQVPL | NPPGTDHVTV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| PLALMFEDVT | VAATNFSFYD | CSAVQALEAA | APCRACVGSI | WRCHWCPQSS | HCVYGEHCPE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| GERTIYSAQE | VDIQVRGPGA | CPQVEGLAGP | HLVPVGWESH | LALRVRNLQH | FRGLPASFHC |
| 730 | 740 | 750 | 760 | 770 | 780 |
| WLELPGELRG | LPATLEETAG | DSGLIHCQAH | QFYPSMSQRE | LPVPIYVTQG | EAQRLDNTHA |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LYVILYDCAM | GHPDCSHCQA | ANRSLGCLWC | ADGQPACRYG | PLCPPGAVEL | LCPAPSIDAV |
| 850 | 860 | 870 | 880 | 890 | 900 |
| EPLTGPPEGG | LALTILGSNL | GRAFADVQYA | VSVASRPCNP | EPSLYRTSAR | IVCVTSPAPN |
| 910 | 920 | 930 | 940 | 950 | 960 |
| GTTGPVRVAI | KSQPPGISSQ | HFTYQDPVLL | SLSPRWGPQA | GGTQLTIRGQ | HLQTGGNTSA |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| FVGGQPCPIL | EPVCPEAIVC | RTRPQAAPGE | AAVLVVFGHA | QRTLLASPFR | YTANPQLVAA |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| EPSASFRGGG | RLIRVRGTGL | DVVQRPLLSV | WLEADAEVQA | SRAQPQDPQP | RRSCGAPAAD |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| PQACIQLGGG | LLQCSTVCSV | NSSSLLLCRS | PAVPDRAHPQ | RVFFTLDNVQ | VDFASASGGQ |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| GFLYQPNPRL | APLSREGPAR | PYRLKPGHVL | DVEGEGLNLG | ISKEEVRVHI | GRGECLVKTL |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| TRTHLYCEPP | AHAPQPANGS | GLPQFVVQMG | NVQLALGPVQ | YEAEPPLSAF | PVEAQAGVGM |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| GAAVLIAAVL | LLTLMYRHKS | KQALRDYQKV | LVQLESLETG | VGDQCRKEFT | DLMTEMTDLS |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| SDLEGSGIPF | LDYRTYAERA | FFPGHGGCPL | QPKPEGPGED | GHCATVRQGL | TQLSNLLNSK |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| LFLLTLIHTL | EEQPSFSQRD | RCHVASLLSL | ALHGKLEYLT | DIMRTLLGDL | AAHYVHRNPK |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| LMLRRTETMV | EKLLTNWLSI | CLYAFLREVA | GEPLYMLFRA | IQYQVDKGPV | DAVTGKAKRT |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| LNDSRLLRED | VEFQPLTLMV | LVGPGAGGAA | GSSEMQRVPA | RVLDTDTITQ | VKEKVLDQVY |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| KGTPFSQRPS | VHALDLEWRS | GLAGHLTLSD | EDLTSVTQNH | WKRLNTLQHY | KVPDGATVGL |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| VPQLHRGSTI | SQSLAQRCPL | GENIPTLEDG | EEGGVCLWHL | VKATEEPEGA | KVRCSSLRER |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| EPARAKAIPE | IYLTRLLSMK | GTLQKFVDDT | FQAILSVNRP | IPIAVKYLFD | LLDELAEKHG |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| IEDPGTLHIW | KTNSLLLRFW | VNALKNPQLI | FDVRVSDNVD | AILAVIAQTF | IDSCTTSEHK |
| 1810 | 1820 | 1830 | 1840 | 1850 | 1860 |
| VGRDSPVNKL | LYAREIPRYK | QMVERYYADI | RQSSPASYQE | MNSALAELSG | NYTSAPHCLE |
| 1870 | 1880 | 1890 | 1900 | ||
| ALQELYNHIH | RYYDQIISAL | EEDPVGQKLQ | LACRLQQVAA | LVENKVTDL |