Q9UHY1
Gene name |
NRBP1 (BCON3) |
Protein name |
Nuclear receptor-binding protein |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:29959 |
EC number |
|
Protein Class |
|
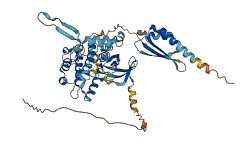
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9UHY1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9UHY1-F1 | Predicted | AlphaFoldDB |
213 variants for Q9UHY1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA44503250 rs919186043 |
2 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA346387349 rs919186043 |
2 | S>W | No |
ClinGen TOPMed gnomAD |
|
|
rs1339699398 CA346387409 |
4 | G>E | No |
ClinGen gnomAD |
|
|
rs1313458435 CA346387400 |
4 | G>R | No |
ClinGen gnomAD |
|
|
CA346387425 rs1279468000 |
5 | E>A | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 5 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1578547 rs771034587 |
5 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA346387448 rs1572687575 |
6 | S>A | No |
ClinGen Ensembl |
|
|
rs1178151171 CA346387475 |
7 | Q>E | No |
ClinGen Ensembl |
|
|
rs774592261 CA1578548 |
7 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA1578550 rs771880350 |
8 | T>R | No |
ClinGen ExAC gnomAD |
|
|
rs775353105 CA1578551 |
9 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1346523771 CA346387572 |
10 | L>R | No |
ClinGen TOPMed gnomAD |
|
|
COSM1019687 CA346387583 rs1212019331 |
11 | S>N | Variant assessed as Somatic; impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs1285308541 CA346387587 |
11 | S>R | No |
ClinGen TOPMed |
|
|
rs1212019331 CA346387582 |
11 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1285959377 CA346387677 |
16 | P>A | No |
ClinGen gnomAD |
|
|
CA346387709 rs1487573545 |
17 | K>N | No |
ClinGen gnomAD |
|
|
CA346387729 rs1572687650 |
18 | V>G | No |
ClinGen Ensembl |
|
|
CA346387750 rs1195945253 |
19 | E>A | No |
ClinGen gnomAD |
|
|
rs760536735 CA1578552 |
21 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1432657667 CA346387807 |
22 | S>P | No |
ClinGen gnomAD |
|
|
rs529123835 CA1578553 |
23 | S>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs951944318 CA44503307 |
24 | A>S | No |
ClinGen TOPMed |
|
|
rs753276316 CA1578554 |
24 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA346387874 rs1478898930 |
26 | G>D | No |
ClinGen gnomAD |
|
|
rs1173987845 CA346387895 |
27 | L>P | No |
ClinGen gnomAD |
|
|
CA346387898 rs1417718063 |
28 | T>A | No |
ClinGen gnomAD |
|
|
CA346387909 rs1384132656 |
28 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1299943305 COSM268000 CA346387951 |
30 | V>A | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs1385700397 CA346388007 |
33 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs755335330 CA1578558 |
37 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1005987885 CA44503336 |
37 | T>I | No |
ClinGen TOPMed |
|
|
rs1572687798 CA346388116 |
38 | T>I | No |
ClinGen Ensembl |
|
|
rs868607849 CA44503342 |
39 | S>L | No |
ClinGen Ensembl |
|
|
CA1578559 rs781693165 |
40 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753170710 CA1578560 |
40 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1313435812 CA346388152 |
41 | A>P | No |
ClinGen gnomAD |
|
|
CA1578561 rs563642807 |
42 | S>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1219509685 CA346388189 |
43 | P>L | No |
ClinGen gnomAD |
|
|
rs1208085042 CA346388298 |
47 | E>D | No |
ClinGen gnomAD |
|
| TCGA novel | 53 | S>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA346388568 rs1379513593 |
63 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA1578579 rs756539212 |
72 | N>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1299031878 CA346388750 |
74 | R>W | No |
ClinGen gnomAD |
|
|
rs764463795 CA1578580 |
81 | S>I | No |
ClinGen ExAC |
|
| TCGA novel | 89 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1335593991 CA346389021 |
94 | V>D | No |
ClinGen gnomAD |
|
|
CA346389018 rs1335593991 |
94 | V>G | No |
ClinGen gnomAD |
|
|
rs199510158 CA44503579 |
101 | F>L | No |
ClinGen Ensembl |
|
|
rs758609544 CA1578585 |
104 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs369322316 CA1578586 |
105 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA346389193 rs1320223026 |
107 | Y>C | No |
ClinGen gnomAD |
|
|
CA44503610 rs11539698 |
109 | L>Q | No |
ClinGen Ensembl |
|
|
rs1558334992 CA346389255 |
111 | E>K | No |
ClinGen Ensembl |
|
|
CA346389432 rs1424001450 |
115 | R>H | No |
ClinGen gnomAD |
|
|
CA1578608 rs781036364 |
138 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1296874066 CA346390068 |
139 | I>V | No |
ClinGen gnomAD |
|
|
rs146226018 CA1578610 |
144 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs778484098 CA1578631 |
147 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA346390786 rs1288457589 |
159 | K>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 162 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA44504116 COSM215874 rs1047817538 |
165 | T>I | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1480678955 COSM1407454 CA346391099 |
171 | T>M | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
rs1416126166 CA346391316 |
177 | W>G | No |
ClinGen gnomAD |
|
|
rs111798412 CA44504261 |
178 | K>R | No |
ClinGen Ensembl |
|
|
CA346391632 rs1235759007 |
194 | C>R | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 195 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs779546002 CA1578670 |
196 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs757867684 CA1578669 |
196 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs983579251 CA44504569 |
197 | P>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1451189311 CA346391685 |
197 | P>S | No |
ClinGen gnomAD |
|
| rs763892200 | 198 | I>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1455394018 CA346391695 |
198 | I>V | No |
ClinGen gnomAD |
|
|
CA44504594 rs1003252519 |
200 | H>R | No |
ClinGen TOPMed |
|
| TCGA novel | 207 | T>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA346391919 rs1284160214 |
220 | S>F | No |
ClinGen gnomAD |
|
|
rs1008624595 CA44507445 |
225 | T>I | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 231 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1159316447 CA346393709 |
233 | C>S | No |
ClinGen TOPMed |
|
|
CA1578701 rs768882955 |
234 | R>* | No |
ClinGen ExAC gnomAD |
|
|
rs776856765 CA1578702 |
234 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs761946471 CA1578703 |
235 | E>K | No |
ClinGen ExAC |
|
|
CA346393909 rs1439238916 |
241 | H>Y | No |
ClinGen TOPMed |
|
|
CA346393942 rs1171482496 |
242 | F>L | No |
ClinGen gnomAD |
|
| TCGA novel | 248 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1578726 rs774202584 |
250 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA44507577 rs201083911 |
261 | S>C | No |
ClinGen Ensembl |
|
|
rs201083911 CA44507582 |
261 | S>F | No |
ClinGen Ensembl |
|
| TCGA novel | 271 | V>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA44507741 rs767730395 |
282 | Y>C | No |
ClinGen Ensembl |
|
|
rs1409910374 CA346394736 |
284 | P>S | No |
ClinGen gnomAD |
|
|
CA346394816 rs1292072778 |
287 | A>P | No |
ClinGen gnomAD |
|
|
CA346394893 rs1353010281 |
290 | S>N | No |
ClinGen gnomAD |
|
|
rs1572691822 CA346394944 |
292 | I>T | No |
ClinGen Ensembl |
|
|
rs147955632 CA1578757 CA1578756 |
293 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA346395107 rs1275723999 |
299 | L>S | No |
ClinGen gnomAD |
|
| TCGA novel | 302 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs71441076 CA44509253 |
310 | S>F | No |
ClinGen Ensembl |
|
|
CA1578781 rs773019385 |
311 | E>D | No |
ClinGen ExAC |
|
|
rs201132320 CA44509268 |
313 | A>V | No |
ClinGen 1000Genomes |
|
|
CA1578783 rs536923313 |
314 | R>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1578782 rs536923313 |
314 | R>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1578784 rs371237665 |
314 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA346396390 rs371237665 |
314 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1243702829 CA346396435 |
316 | P>S | No |
ClinGen gnomAD |
|
|
CA1578786 rs764631733 |
320 | E>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 324 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1357154706 CA346396641 |
325 | P>L | No |
ClinGen TOPMed |
|
|
CA1578790 rs750674505 |
333 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA1578791 rs758065640 |
337 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1578794 rs145354088 |
341 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs371384227 CA1578796 |
344 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1578798 rs772672860 |
346 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758633337 CA1578809 |
347 | M>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 350 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1229817734 CA346397586 |
352 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 353 | L>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
TCGA novel rs754853866 CA1578812 |
355 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
NCI-TCGA ClinGen ExAC gnomAD |
|
rs368054308 CA346397705 |
356 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1433522225 CA346397806 |
360 | M>V | No |
ClinGen TOPMed |
|
|
CA346397866 rs1474704385 |
361 | D>N | No |
ClinGen gnomAD |
|
|
CA44509717 rs769914699 |
362 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1578815 rs769914699 |
362 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
RCV000967947 rs56004639 VAR_041359 CA1578818 |
365 | V>I | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1393243890 CA346398194 |
370 | P>R | No |
ClinGen gnomAD |
|
|
rs1411755765 CA346398199 |
371 | A>T | No |
ClinGen TOPMed |
|
|
rs773905346 CA1578819 |
375 | R>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 376 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs777057653 CA1578822 |
378 | V>F | No |
ClinGen ExAC gnomAD |
|
|
rs1482491959 CA346398647 |
381 | L>W | No |
ClinGen gnomAD |
|
|
CA346399412 rs1477664839 |
383 | S>Y | No |
ClinGen TOPMed |
|
| TCGA novel | 393 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1578858 rs369958327 |
399 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs779726099 CA1578859 |
400 | G>E | No |
ClinGen ExAC gnomAD |
|
|
rs748475059 CA1578860 |
402 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA346399931 rs1193397805 |
402 | Y>H | No |
ClinGen gnomAD |
|
|
CA1578861 rs770079968 |
406 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1157212067 CA346400008 |
407 | F>S | No |
ClinGen gnomAD |
|
|
CA346400034 rs1391019223 |
409 | L>P | No |
ClinGen gnomAD |
|
|
CA346400046 rs1458643486 |
410 | P>L | No |
ClinGen gnomAD |
|
|
CA1578864 rs372579058 |
411 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM1019689 CA1578863 rs372579058 |
411 | R>Q | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs199714909 CA1578862 |
411 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 413 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA346400085 COSM1019690 rs774546607 |
413 | Q>H | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA44509943 rs11539699 |
414 | Q>* | No |
ClinGen Ensembl |
|
|
rs1572696364 CA346400214 |
420 | V>A | No |
ClinGen Ensembl |
|
|
CA346400274 rs1465315760 |
424 | V>F | No |
ClinGen TOPMed |
|
|
CA346400272 rs1465315760 |
424 | V>I | No |
ClinGen TOPMed |
|
|
rs767995482 CA1578867 |
425 | V>M | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 426 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1330713982 CA346400308 |
426 | P>L | No |
ClinGen gnomAD |
|
|
rs760459804 CA346400321 |
427 | P>H | No |
ClinGen ExAC gnomAD |
|
|
CA1578869 rs760459804 |
427 | P>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 428 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs763804268 CA1578870 |
428 | S>P | No |
ClinGen ExAC gnomAD |
|
|
rs1410562987 CA346400378 |
431 | T>S | No |
ClinGen TOPMed |
|
|
rs753734841 VAR_041360 COSM20720 CA1578872 |
432 | P>L | ovary an ovarian mucinous carcinoma sample; somatic mutation [Cosmic, UniProt] | No |
ClinGen cosmic curated UniProt ExAC dbSNP gnomAD |
|
CA1578873 rs761644081 |
433 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1180949430 CA346400461 |
436 | P>Q | No |
ClinGen TOPMed |
|
|
CA1578874 rs765209087 |
442 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs1226923253 CA346400852 |
448 | Q>L | No |
ClinGen gnomAD |
|
|
rs574325125 CA1578896 |
453 | S>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA1578899 rs754561047 |
455 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs751134996 CA1578898 |
455 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs34260196 CA346401263 |
460 | H>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs34260196 VAR_041361 CA1578901 |
460 | H>R | No |
ClinGen UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs138754846 | 461 | H>= | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs199938360 CA1578902 |
461 | H>Y | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 462 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA346401439 rs1186395768 |
463 | T>K | No |
ClinGen gnomAD |
|
|
rs1308187702 CA346401453 |
464 | L>V | No |
ClinGen TOPMed |
|
|
rs1450060996 CA346401470 |
465 | L>M | No |
ClinGen gnomAD |
|
|
rs773070005 CA1578914 |
471 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs546007305 CA346401702 |
472 | L>V | No |
ClinGen 1000Genomes gnomAD |
|
|
rs773699029 CA1578917 |
474 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767027817 CA1578919 |
475 | H>N | No |
ClinGen ExAC gnomAD |
|
|
rs765694579 CA1578922 |
482 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs750877807 CA1578923 |
483 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs750877807 CA44510327 |
483 | N>H | No |
ClinGen ExAC gnomAD |
|
|
CA44510609 rs977150998 |
485 | N>H | No |
ClinGen Ensembl |
|
|
rs1228525765 CA346402393 |
487 | P>L | No |
ClinGen TOPMed |
|
|
rs781330388 CA1578950 |
488 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1578951 rs374507004 |
490 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs754046270 CA44510625 |
499 | I>V | No |
ClinGen Ensembl |
|
|
rs756295007 CA1578952 |
500 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA1578974 rs757484710 |
504 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA44510738 rs141808993 |
504 | Q>R | No |
ClinGen ESP |
|
|
rs1381091916 CA346402884 |
505 | S>I | No |
ClinGen gnomAD |
|
|
CA346402881 rs1381091916 |
505 | S>N | No |
ClinGen gnomAD |
|
|
rs778753969 CA1578976 |
506 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA346402891 rs141700147 |
506 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1578977 COSM184422 rs141700147 |
506 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs778753969 CA1578975 |
506 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA346402932 rs1362882652 |
508 | T>I | No |
ClinGen gnomAD |
|
|
CA1578978 rs780172704 |
510 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA1578979 rs746470640 |
516 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs866974645 CA44510784 |
516 | N>S | No |
ClinGen Ensembl |
|
|
rs1483784394 CA346403199 |
517 | K>N | No |
ClinGen gnomAD |
|
|
CA346403209 rs1208451074 |
518 | F>I | No |
ClinGen gnomAD |
|
|
rs768298741 CA1578980 |
519 | N>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
COSM418753 CA346403323 rs1451925011 |
521 | A>V | urinary_tract [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA44510798 rs1003536519 |
522 | R>S | No |
ClinGen Ensembl |
|
|
rs1025675347 CA44510811 |
523 | N>D | No |
ClinGen Ensembl |
|
| TCGA novel | 525 | T>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1578981 rs371589798 |
525 | T>I | No |
ClinGen ESP ExAC gnomAD |
|
|
CA346403405 rs1572698259 |
525 | T>P | No |
ClinGen Ensembl |
|
|
rs1427251074 CA346403427 |
526 | L>F | No |
ClinGen TOPMed |
|
|
rs761556440 CA1578982 |
527 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs887157695 CA44510818 |
528 | S>L | No |
ClinGen TOPMed gnomAD |
|
|
CA1578983 rs769393462 |
530 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1437274482 CA346403660 |
533 | V>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
No associated diseases with Q9UHY1
3 regional properties for Q9UHY1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Heat shock protein 70, conserved site | 9 - 16 | IPR018181-1 |
| conserved_site | Heat shock protein 70, conserved site | 197 - 210 | IPR018181-2 |
| conserved_site | Heat shock protein 70, conserved site | 334 - 348 | IPR018181-3 |
Functions
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| endomembrane system | A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| endoplasmic reticulum to Golgi vesicle-mediated transport | The directed movement of substances from the endoplasmic reticulum (ER) to the Golgi, mediated by COP II vesicles. Small COP II coated vesicles form from the ER and then fuse directly with the cis-Golgi. Larger structures are transported along microtubules to the cis-Golgi. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P51957 | NEK4 | Serine/threonine-protein kinase Nek4 | Homo sapiens (Human) | PR |
| Q9Y3S1 | WNK2 | Serine/threonine-protein kinase WNK2 | Homo sapiens (Human) | SS |
| Q9H4A3 | WNK1 | Serine/threonine-protein kinase WNK1 | Homo sapiens (Human) | SS |
| Q9BYP7 | WNK3 | Serine/threonine-protein kinase WNK3 | Homo sapiens (Human) | SS |
| Q9Y2U5 | MAP3K2 | Mitogen-activated protein kinase kinase kinase 2 | Homo sapiens (Human) | PR |
| Q99759 | MAP3K3 | Mitogen-activated protein kinase kinase kinase 3 | Homo sapiens (Human) | PR |
| Q99J45 | Nrbp1 | Nuclear receptor-binding protein | Mus musculus (Mouse) | PR |
| Q9STK6 | WNK3 | Probable serine/threonine-protein kinase WNK3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RXE5 | WNK10 | Probable serine/threonine-protein kinase WNK10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q944Q0 | WNK8 | Serine/threonine-protein kinase WNK8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LVL5 | WNK4 | Probable serine/threonine-protein kinase WNK4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LST2 | WNK7 | Probable serine/threonine-protein kinase WNK7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSEGESQTVL | SSGSDPKVES | SSSAPGLTSV | SPPVTSTTSA | ASPEEEEESE | DESEILEESP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CGRWQKRREE | VNQRNVPGID | SAYLAMDTEE | GVEVVWNEVQ | FSERKNYKLQ | EEKVRAVFDN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LIQLEHLNIV | KFHKYWADIK | ENKARVIFIT | EYMSSGSLKQ | FLKKTKKNHK | TMNEKAWKRW |
| 190 | 200 | 210 | 220 | 230 | 240 |
| CTQILSALSY | LHSCDPPIIH | GNLTCDTIFI | QHNGLIKIGS | VAPDTINNHV | KTCREEQKNL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| HFFAPEYGEV | TNVTTAVDIY | SFGMCALEMA | VLEIQGNGES | SYVPQEAISS | AIQLLEDPLQ |
| 310 | 320 | 330 | 340 | 350 | 360 |
| REFIQKCLQS | EPARRPTARE | LLFHPALFEV | PSLKLLAAHC | IVGHQHMIPE | NALEEITKNM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| DTSAVLAEIP | AGPGREPVQT | LYSQSPALEL | DKFLEDVRNG | IYPLTAFGLP | RPQQPQQEEV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TSPVVPPSVK | TPTPEPAEVE | TRKVVLMQCN | IESVEEGVKH | HLTLLLKLED | KLNRHLSCDL |
| 490 | 500 | 510 | 520 | 530 | |
| MPNENIPELA | AELVQLGFIS | EADQSRLTSL | LEETLNKFNF | ARNSTLNSAA | VTVSS |