Q9T014
Gene name |
SPA2 (At4g11110, F2P3.13, F2P3.14, T22B4.90) |
Protein name |
Protein SPA1-RELATED 2 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT4G11110 |
EC number |
|
Protein Class |
|
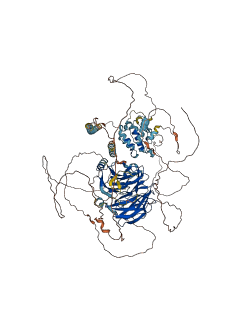
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9T014
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9T014-F1 | Predicted | AlphaFoldDB |
92 variants for Q9T014
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH00489245 | 5 | G>R | No | 1000Genomes | |
| ENSVATH11487263 | 8 | G>A | No | 1000Genomes | |
| tmp_4_6772226_C_A | 22 | Q>K | No | 1000Genomes | |
| ENSVATH00489246 | 28 | Q>H | No | 1000Genomes | |
| ENSVATH06613163 | 29 | S>P | No | 1000Genomes | |
| ENSVATH06613164 | 32 | P>L | No | 1000Genomes | |
| ENSVATH06613164 | 32 | P>R | No | 1000Genomes | |
| ENSVATH00489248 | 89 | E>A | No | 1000Genomes | |
| ENSVATH06613167 | 92 | V>F | No | 1000Genomes | |
| ENSVATH06613168 | 116 | A>P | No | 1000Genomes | |
| tmp_4_6772526_C_T | 122 | R>C | No | 1000Genomes | |
| ENSVATH14131818 | 136 | P>Q | No | 1000Genomes | |
| tmp_4_6772587_G_T | 142 | S>I | No | 1000Genomes | |
| ENSVATH06613169 | 146 | I>T | No | 1000Genomes | |
| ENSVATH06613170 | 156 | N>T | No | 1000Genomes | |
| ENSVATH06613171 | 159 | K>N | No | 1000Genomes | |
| tmp_4_6772640_A_T | 160 | M>L | No | 1000Genomes | |
| tmp_4_6772674_C_A | 171 | A>D | No | 1000Genomes | |
| tmp_4_6772692_G_A | 177 | G>E | No | 1000Genomes | |
| ENSVATH06613172 | 180 | N>S | No | 1000Genomes | |
| tmp_4_6772719_T_C | 186 | V>A | No | 1000Genomes | |
| ENSVATH11487295 | 188 | R>Q | No | 1000Genomes | |
| ENSVATH00489249 | 198 | H>Y | No | 1000Genomes | |
| tmp_4_6772763_A_T | 201 | I>F | No | 1000Genomes | |
| ENSVATH00489251 | 251 | A>D | No | 1000Genomes | |
| ENSVATH11487297 | 254 | I>V | No | 1000Genomes | |
| tmp_4_6772938_C_T | 259 | A>V | No | 1000Genomes | |
| tmp_4_6772950_G_C | 263 | S>T | No | 1000Genomes | |
| ENSVATH11487298 | 268 | A>E | No | 1000Genomes | |
| ENSVATH11487298 | 268 | A>V | No | 1000Genomes | |
| tmp_4_6772970_T_A | 270 | Y>N | No | 1000Genomes | |
| ENSVATH06613174 | 273 | L>F | No | 1000Genomes | |
| tmp_4_6772983_C_G | 274 | P>R | No | 1000Genomes | |
| tmp_4_6773001_C_T | 280 | T>I | No | 1000Genomes | |
| ENSVATH06613175 | 292 | T>S | No | 1000Genomes | |
| tmp_4_6773040_A_G | 293 | H>R | No | 1000Genomes | |
| tmp_4_6773091_A_G | 310 | E>G | No | 1000Genomes | |
| tmp_4_6773098_G_T | 312 | Q>H | No | 1000Genomes | |
| ENSVATH02784243 | 327 | V>M | No | 1000Genomes | |
| tmp_4_6773156_T_C | 332 | C>R | No | 1000Genomes | |
| ENSVATH06613176 | 334 | H>L | No | 1000Genomes | |
| tmp_4_6773193_G_A | 344 | R>Q | No | 1000Genomes | |
| tmp_4_6773198_T_A | 346 | S>T | No | 1000Genomes | |
| tmp_4_6773207_A_G | 349 | K>E | No | 1000Genomes | |
| ENSVATH06613177 | 353 | E>V | No | 1000Genomes | |
| ENSVATH14131819 | 354 | N>D | No | 1000Genomes | |
| tmp_4_6773325_G_T | 388 | R>L | No | 1000Genomes | |
| tmp_4_6773351_A_G | 397 | I>V | No | 1000Genomes | |
| tmp_4_6773424_T_A | 421 | V>D | No | 1000Genomes | |
| ENSVATH06613178 | 434 | E>A | No | 1000Genomes | |
| tmp_4_6773507_T_C | 449 | C>R | No | 1000Genomes | |
| tmp_4_6773975_C_G | 493 | Q>E | No | 1000Genomes | |
| ENSVATH11487347 | 501 | E>D | No | 1000Genomes | |
| ENSVATH02784246 | 546 | Q>H | No | 1000Genomes | |
| ENSVATH06613194 | 554 | P>S | No | 1000Genomes | |
| tmp_4_6774541_G_C | 556 | L>F | No | 1000Genomes | |
| ENSVATH02784247 | 566 | E>D | No | 1000Genomes | |
| ENSVATH11487354 | 579 | L>V | No | 1000Genomes | |
| tmp_4_6774645_G_C | 591 | G>A | No | 1000Genomes | |
| ENSVATH02784248 | 594 | M>I | No | 1000Genomes | |
| ENSVATH02784249 | 595 | E>K | No | 1000Genomes | |
| ENSVATH02784250 | 596 | E>Q | No | 1000Genomes | |
| ENSVATH11487365 | 598 | A>E | No | 1000Genomes | |
| ENSVATH11487365 | 598 | A>V | No | 1000Genomes | |
| tmp_4_6774686_G_A | 605 | E>K | No | 1000Genomes | |
| tmp_4_6774750_C_A | 626 | S>Y | No | 1000Genomes | |
| ENSVATH00489261 | 645 | S>C | No | 1000Genomes | |
| ENSVATH02784251 | 645 | S>R | No | 1000Genomes | |
| tmp_4_6774810_C_T | 646 | A>V | No | 1000Genomes | |
| tmp_4_6774833_G_A | 654 | A>T | No | 1000Genomes | |
| tmp_4_6774837_A_G | 655 | H>R | No | 1000Genomes | |
| ENSVATH00489262 | 659 | A>T | No | 1000Genomes | |
| tmp_4_6774871_T_A | 666 | D>E | No | 1000Genomes | |
| ENSVATH11487368 | 670 | L>Q | No | 1000Genomes | |
| ENSVATH06613196 | 672 | N>D | No | 1000Genomes | |
| tmp_4_6774893_G_A | 674 | D>N | No | 1000Genomes | |
| ENSVATH06613197 | 679 | E>K | No | 1000Genomes | |
| tmp_4_6774980_G_T | 703 | A>S | No | 1000Genomes | |
| ENSVATH06613199 | 717 | S>I | No | 1000Genomes | |
| tmp_4_6775028_T_G | 719 | L>V | No | 1000Genomes | |
| ENSVATH06613201 | 732 | D>N | No | 1000Genomes | |
| ENSVATH11487373 | 733 | R>Q | No | 1000Genomes | |
| ENSVATH00489265 | 755 | L>V | No | 1000Genomes | |
| tmp_4_6775180_G_A | 769 | M>I | No | 1000Genomes | |
| tmp_4_6775486_G_A | 838 | C>Y | No | 1000Genomes | |
| ENSVATH02784253 | 868 | Q>H | No | 1000Genomes | |
| ENSVATH11487406 | 912 | N>T | No | 1000Genomes | |
| ENSVATH06613212 | 965 | I>V | No | 1000Genomes | |
| ENSVATH06613220 | 975 | Y>F | No | 1000Genomes | |
| tmp_4_6776503_C_G | 980 | S>C | No | 1000Genomes | |
| tmp_4_6776610_A_C | 1016 | K>Q | No | 1000Genomes | |
| ENSVATH08326223 | 1022 | V>A | No | 1000Genomes |
No associated diseases with Q9T014
12 regional properties for Q9T014
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 200 - 550 | IPR000719 |
| repeat | WD40 repeat | 712 - 751 | IPR001680-1 |
| repeat | WD40 repeat | 762 - 886 | IPR001680-2 |
| repeat | WD40 repeat | 890 - 930 | IPR001680-3 |
| repeat | WD40 repeat | 939 - 978 | IPR001680-4 |
| repeat | WD40 repeat | 994 - 1034 | IPR001680-5 |
| conserved_site | WD40 repeat, conserved site | 788 - 802 | IPR019775-1 |
| conserved_site | WD40 repeat, conserved site | 831 - 845 | IPR019775-2 |
| conserved_site | WD40 repeat, conserved site | 915 - 929 | IPR019775-3 |
| repeat | G-protein beta WD-40 repeat | 788 - 802 | IPR020472-1 |
| repeat | G-protein beta WD-40 repeat | 831 - 845 | IPR020472-2 |
| repeat | G-protein beta WD-40 repeat | 915 - 929 | IPR020472-3 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| Cul4-RING E3 ubiquitin ligase complex | A ubiquitin ligase complex in which a cullin from the Cul4 family and a RING domain protein form the catalytic core; substrate specificity is conferred by an adaptor protein. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| photomorphogenesis | The control of plant growth, development, and differentiation by the duration and nature of light, independent of photosynthesis. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| red, far-red light phototransduction | The sequence of reactions within a cell required to convert absorbed photons from red or far-red light into a molecular signal; the red, far-red light range is defined as having a wavelength within the range 660-730 nm. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P19525 | EIF2AK2 | Interferon-induced, double-stranded RNA-activated protein kinase | Homo sapiens (Human) | PR |
| Q9NZJ5 | EIF2AK3 | Eukaryotic translation initiation factor 2-alpha kinase 3 | Homo sapiens (Human) | PR |
| Q8NHY2 | COP1 | E3 ubiquitin-protein ligase COP1 | Homo sapiens (Human) | PR |
| Q9R1A8 | Cop1 | E3 ubiquitin-protein ligase COP1 | Mus musculus (Mouse) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MMDEGSVGDV | SRIDEADVAH | LQFKNSEQSF | KPENIEVREV | KEVQVQREAG | SPDCSYGVIA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DFLDGKNGGD | HVELIGNEPC | SSRQNTNDEG | DVVEELTVKT | CEGSSMAIVG | RPSSRARLEM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NRSQFLHRFP | LDGDLPGSSS | MSKKVIDRGT | VSILRNAGKM | SLPETSNGQL | AIIAVNGEAN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EHLTNVERNP | VPVEALSHEG | IKTKMLSQSG | FSQFFVRKTL | KGKGVTFRGP | PNNRSKARNM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DQQTVASSGS | ALVIANTSAK | ISSSIPLAAY | DGLPCLPSNT | SKPSSCANPS | DTHRGCGGEG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LSLREWLKSE | RQEVNKAECM | YIFRQIVDHV | DCSHSQGVVL | CDLRPSSFKI | FKENAVKYVV |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SGSQRESFDS | NMNKETLSQL | ENPLVRRRLG | DTSSLSIPAK | KQKSSGPSSR | QWPMFQRAGG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VNIQTENNDG | AIQEFHFRSS | QPHCSTVACP | FTSVSEQLEE | KWYASPEELR | GDMRSASSNI |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YSLGILLYEL | LSQFQCERAR | EAAMSDIRHR | ILPPKFLSEN | PKEAGFCLWL | LHPESSCRPS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| TRDILQSEVV | NGIPDLYAEG | LSLSIEQEDT | ESELLQHFLF | LSQEKRQKHA | GNLMEEIASV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EADIEEIVKR | RCAIGPPSLE | EASSSSPASS | VPEMRLIRNI | NQLESAYFAA | RIDAHLPEAR |
| 670 | 680 | 690 | 700 | 710 | 720 |
| YRLRPDRDLL | RNSDNTVAEV | ENSETWSSDD | RVGAFFDGLC | KYARYSKFET | RGVLRTSELN |
| 730 | 740 | 750 | 760 | 770 | 780 |
| NTSNVICSLG | FDRDEDYFAT | AGVSKKIKIY | EFNSLFNESV | DIHYPAIEMP | NRSKLSGVCW |
| 790 | 800 | 810 | 820 | 830 | 840 |
| NNYIRNYLAS | SDYDGIVKLW | DVTTGQAISH | FIEHEKRAWS | VDFSEACPTK | LASGSDDCSV |
| 850 | 860 | 870 | 880 | 890 | 900 |
| KLWNINERNC | LGTIRNIANV | CCVQFSPQSS | HLLAFGSSDF | RTYCYDLRNL | RTPWCILSGH |
| 910 | 920 | 930 | 940 | 950 | 960 |
| NKAVSYAKFL | DNETLVTAST | DNTLKLWDLK | KTTHGGLSTN | ACSLTFGGHT | NEKNFVGLST |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| SDGYIACGSE | TNEVYAYHRS | LPMPITSYKF | GSIDPISGKE | IEEDNNLFVS | SVCWRKRSNM |
| 1030 | |||||
| VVSASSNGSI | KVLQLV |