Q9SNA3
Gene name |
At3g46340 (F18L15.60) |
Protein name |
Putative receptor-like protein kinase At3g46340 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G46340 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
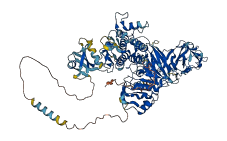
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
728-735 (Activation loop from InterPro)
Target domain |
591-857 (Catalytic domain of the Serine/Threonine kinases, Interleukin-1 Receptor Associated Kinases and related STKs) |
Relief mechanism |
|
Assay |
|
746-753 (Activation loop from InterPro)
Target domain |
591-857 (Catalytic domain of the Serine/Threonine kinases, Interleukin-1 Receptor Associated Kinases and related STKs) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9SNA3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9SNA3-F1 | Predicted | AlphaFoldDB |
154 variants for Q9SNA3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH06245254 | 8 | L>V | No | 1000Genomes | |
| ENSVATH14380683 | 15 | A>E | No | 1000Genomes | |
| ENSVATH06245281 | 68 | I>L | No | 1000Genomes | |
| ENSVATH06245281 | 68 | I>V | No | 1000Genomes | |
| ENSVATH14380741 | 69 | G>S | No | 1000Genomes | |
| ENSVATH06245283 | 79 | T>P | No | 1000Genomes | |
| ENSVATH14380742 | 97 | Y>* | No | 1000Genomes | |
| ENSVATH00402363 | 105 | K>T | No | 1000Genomes | |
| tmp_3_17028188_C_A | 114 | L>I | No | 1000Genomes | |
| tmp_3_17028189_T_C | 114 | L>P | No | 1000Genomes | |
| tmp_3_17028191_T_A | 115 | Y>N | No | 1000Genomes | |
| ENSVATH00402364 | 121 | L>H | No | 1000Genomes | |
| ENSVATH00402364 | 121 | L>R | No | 1000Genomes | |
| tmp_3_17028252_T_A | 135 | F>Y | No | 1000Genomes | |
| tmp_3_17028258_C_T | 137 | T>I | No | 1000Genomes | |
| ENSVATH06245285 | 143 | E>G | No | 1000Genomes | |
| ENSVATH06245284 | 143 | E>K | No | 1000Genomes | |
| ENSVATH06245286 | 146 | S>T | No | 1000Genomes | |
| tmp_3_17028297_A_C | 150 | E>A | No | 1000Genomes | |
| ENSVATH06245287 | 158 | S>* | No | 1000Genomes | |
| ENSVATH02448168 | 168 | T>K | No | 1000Genomes | |
| tmp_3_17028357_C_A | 170 | T>K | No | 1000Genomes | |
| tmp_3_17028372_T_C | 175 | L>S | No | 1000Genomes | |
| tmp_3_17028393_C_T | 182 | P>L | No | 1000Genomes | |
| tmp_3_17028417_C_G | 190 | T>S | No | 1000Genomes | |
| ENSVATH06245288 | 191 | G>D | No | 1000Genomes | |
| ENSVATH14380744 | 197 | T>S | No | 1000Genomes | |
| ENSVATH06245290 | 199 | R>G | No | 1000Genomes | |
| tmp_3_17028445_G_C | 199 | R>S | No | 1000Genomes | |
| ENSVATH14380746 | 216 | V>F | No | 1000Genomes | |
| ENSVATH06245295 | 227 | S>T | No | 1000Genomes | |
| ENSVATH06245297 | 233 | W>C | No | 1000Genomes | |
| ENSVATH02448174 | 233 | W>S | No | 1000Genomes | |
| ENSVATH02448175 | 237 | K>R | No | 1000Genomes | |
| ENSVATH02448176 | 243 | G>R | No | 1000Genomes | |
| ENSVATH02448176 | 243 | G>S | No | 1000Genomes | |
| tmp_3_17028662_T_G | 245 | L>R | No | 1000Genomes | |
| ENSVATH02448177 | 245 | L>V | No | 1000Genomes | |
| tmp_3_17028664_G_C | 246 | V>L | No | 1000Genomes | |
| ENSVATH06245299 | 248 | K>Q | No | 1000Genomes | |
| tmp_3_17028671_A_C | 248 | K>T | No | 1000Genomes | |
| ENSVATH02448179 | 249 | N>I | No | 1000Genomes | |
| ENSVATH02448178 | 249 | N>Y | No | 1000Genomes | |
| tmp_3_17028682_A_T | 252 | M>L | No | 1000Genomes | |
| tmp_3_17028720_T_A | 264 | F>L | No | 1000Genomes | |
| ENSVATH06245300 | 265 | R>G | No | 1000Genomes | |
| ENSVATH02448180 | 271 | D>N | No | 1000Genomes | |
| tmp_3_17028778_T_G | 284 | S>A | No | 1000Genomes | |
| ENSVATH12561738 | 296 | E>D | No | 1000Genomes | |
| tmp_3_17028860_TCC_GCC,T | 311 | I>S | No | 1000Genomes | |
| ENSVATH06245302 | 325 | P>L | No | 1000Genomes | |
| ENSVATH02448184 | 326 | V>F | No | 1000Genomes | |
| ENSVATH12561740 | 344 | T>S | No | 1000Genomes | |
| tmp_3_17029000_G_A | 358 | V>I | No | 1000Genomes | |
| ENSVATH12561744 | 361 | P>S | No | 1000Genomes | |
| ENSVATH02448188 | 375 | D>G | No | 1000Genomes | |
| ENSVATH02448187 | 375 | D>N | No | 1000Genomes | |
| ENSVATH02448190 | 379 | T>N | No | 1000Genomes | |
| tmp_3_17029177_C_G | 388 | Q>E | No | 1000Genomes | |
| ENSVATH02448192 | 400 | E>D | No | 1000Genomes | |
| ENSVATH02448193 | 409 | A>T | No | 1000Genomes | |
| ENSVATH06245307 | 414 | R>K | No | 1000Genomes | |
| ENSVATH12561761 | 416 | T>I | No | 1000Genomes | |
| ENSVATH12561763 | 419 | N>S | No | 1000Genomes | |
| tmp_3_17029364_T_A | 421 | S>T | No | 1000Genomes | |
| tmp_3_17029376_T_A | 425 | L>I | No | 1000Genomes | |
| ENSVATH02448199 | 439 | L>V | No | 1000Genomes | |
| ENSVATH02448200 | 440 | D>E | No | 1000Genomes | |
| ENSVATH02448201 | 441 | K>R | No | 1000Genomes | |
| ENSVATH06245311 | 453 | V>L | No | 1000Genomes | |
| ENSVATH06245312 | 459 | S>G | No | 1000Genomes | |
| ENSVATH02448206 | 459 | S>N | No | 1000Genomes | |
| ENSVATH06245319 | 467 | N>K | No | 1000Genomes | |
| tmp_3_17029672_C_A | 471 | N>K | No | 1000Genomes | |
| ENSVATH02448214 | 479 | Q>L | No | 1000Genomes | |
| ENSVATH02448215 | 480 | A>G | No | 1000Genomes | |
| ENSVATH14380747 | 487 | D>N | No | 1000Genomes | |
| ENSVATH02448219 | 496 | Q>E | No | 1000Genomes | |
| tmp_3_17029853_G_C | 502 | G>R | No | 1000Genomes | |
| tmp_3_17029890_T_C | 514 | M>T | No | 1000Genomes | |
| ENSVATH06245324 | 518 | L>* | No | 1000Genomes | |
| ENSVATH02448223 | 526 | I>V | No | 1000Genomes | |
| ENSVATH12561832 | 531 | V>I | No | 1000Genomes | |
| ENSVATH12561833 | 532 | L>F | No | 1000Genomes | |
| ENSVATH02448225 | 540 | K>N | No | 1000Genomes | |
| ENSVATH06245326 | 540 | K>T | No | 1000Genomes | |
| ENSVATH06245327 | 541 | P>L | No | 1000Genomes | |
| ENSVATH02448226 | 545 | E>K | No | 1000Genomes | |
| ENSVATH06245328 | 547 | L>Q | No | 1000Genomes | |
| ENSVATH14380748 | 556 | E>D | No | 1000Genomes | |
| ENSVATH06245329 | 558 | I>V | No | 1000Genomes | |
| ENSVATH12561853 | 559 | T>A | No | 1000Genomes | |
| ENSVATH02448233 | 561 | T>I | No | 1000Genomes | |
| ENSVATH06245330 | 569 | E>K | No | 1000Genomes | |
| ENSVATH12561854 | 576 | S>T | No | 1000Genomes | |
| ENSVATH02448237 | 578 | S>L | No | 1000Genomes | |
| ENSVATH02448239 | 585 | K>N | No | 1000Genomes | |
| ENSVATH06245331 | 588 | Q>L | No | 1000Genomes | |
| tmp_3_17030196_C_A | 590 | P>T | No | 1000Genomes | |
| ENSVATH14380750 | 599 | V>I | No | 1000Genomes | |
| ENSVATH06245332 | 610 | Q>E | No | 1000Genomes | |
| ENSVATH06245333 | 610 | Q>R | No | 1000Genomes | |
| ENSVATH06245334 | 613 | V>I | No | 1000Genomes | |
| ENSVATH06245341 | 649 | C>W | No | 1000Genomes | |
| tmp_3_17030466_C_T | 652 | R>* | No | 1000Genomes | |
| tmp_3_17030469_G_C | 653 | D>H | No | 1000Genomes | |
| ENSVATH12561882 | 661 | Y>S | No | 1000Genomes | |
| ENSVATH02448254 | 671 | L>M | No | 1000Genomes | |
| ENSVATH06245350 | 675 | H>D | No | 1000Genomes | |
| tmp_3_17030625_T_C | 678 | S>P | No | 1000Genomes | |
| ENSVATH12561916 | 679 | V>I | No | 1000Genomes | |
| ENSVATH06245352 | 687 | Q>E | No | 1000Genomes | |
| ENSVATH12561917 | 692 | A>S | No | 1000Genomes | |
| tmp_3_17030668_C_T | 692 | A>V | No | 1000Genomes | |
| ENSVATH06245381 | 722 | Q>H | No | 1000Genomes | |
| ENSVATH06245382 | 724 | T>I | No | 1000Genomes | |
| ENSVATH02448270 | 727 | M>I | No | 1000Genomes | |
| ENSVATH06245383 | 728 | A>V | No | 1000Genomes | |
| ENSVATH06245384 | 734 | R>K | No | 1000Genomes | |
| tmp_3_17031187_C_T | 735 | S>F | No | 1000Genomes | |
| ENSVATH12561959 | 736 | F>S | No | 1000Genomes | |
| ENSVATH00402378 | 743 | Q>H | No | 1000Genomes | |
| ENSVATH12561960 | 747 | V>I | No | 1000Genomes | |
| tmp_3_17031228_G_A | 749 | A>T | No | 1000Genomes | |
| tmp_3_17031240_G_A | 753 | G>R | No | 1000Genomes | |
| tmp_3_17031454_G_A | 761 | R>K | No | 1000Genomes | |
| tmp_3_17031468_G_C | 766 | A>P | No | 1000Genomes | |
| tmp_3_17031476_G_A | 768 | M>I | No | 1000Genomes | |
| tmp_3_17031475_T_C | 768 | M>T | No | 1000Genomes | |
| ENSVATH06245390 | 782 | I>M | No | 1000Genomes | |
| ENSVATH02448294 | 790 | P>Q | No | 1000Genomes | |
| ENSVATH00402382 | 791 | A>T | No | 1000Genomes | |
| tmp_3_17031552_A_T | 794 | K>* | No | 1000Genomes | |
| ENSVATH08141402 | 804 | M>I | No | 1000Genomes | |
| tmp_3_17031618_C_A | 816 | P>T | No | 1000Genomes | |
| ENSVATH06245394 | 817 | N>K | No | 1000Genomes | |
| ENSVATH09433264 | 819 | Q>H | No | 1000Genomes | |
| ENSVATH14380802 | 819 | Q>K | No | 1000Genomes | |
| ENSVATH12561964 | 819 | Q>R | No | 1000Genomes | |
| tmp_3_17031637_A_C | 822 | Y>S | No | 1000Genomes | |
| tmp_3_17031652_T_G | 827 | V>G | No | 1000Genomes | |
| ENSVATH12562005 | 828 | W>* | No | 1000Genomes | |
| ENSVATH02448297 | 835 | M>L | No | 1000Genomes | |
| ENSVATH06245396 | 841 | S>L | No | 1000Genomes | |
| tmp_3_17031699_G_C | 843 | E>Q | No | 1000Genomes | |
| ENSVATH06245397 | 844 | K>N | No | 1000Genomes | |
| tmp_3_17031712_G_C | 847 | S>T | No | 1000Genomes | |
| ENSVATH12562006 | 859 | I>M | No | 1000Genomes | |
| tmp_3_17031768_C_G | 866 | Q>E | No | 1000Genomes | |
| ENSVATH02448303 | 874 | F>L | No | 1000Genomes | |
| tmp_3_17031797_A_C | 875 | E>D | No | 1000Genomes | |
| ENSVATH06245398 | 877 | S>T | No | 1000Genomes | |
| ENSVATH06245399 | 881 | D>E | No | 1000Genomes | |
| ENSVATH14380803 | 884 | A>S | No | 1000Genomes |
No associated diseases with Q9SNA3
3 regional properties for Q9SNA3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Zinc finger C2H2-type | 272 - 301 | IPR013087-1 |
| domain | Zinc finger C2H2-type | 302 - 331 | IPR013087-2 |
| domain | Zinc finger C2H2-type | 332 - 355 | IPR013087-3 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
36 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q91044 | NTRK3 | NT-3 growth factor receptor | Gallus gallus (Chicken) | SS |
| Q91987 | NTRK2 | BDNF/NT-3 growth factors receptor | Gallus gallus (Chicken) | SS |
| Q91009 | NTRK1 | High affinity nerve growth factor receptor | Gallus gallus (Chicken) | SS |
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| Q07407 | htl | Fibroblast growth factor receptor homolog 1 | Drosophila melanogaster (Fruit fly) | PR |
| P20594 | NPR2 | Atrial natriuretic peptide receptor 2 | Homo sapiens (Human) | PR |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P24786 | NTRK3 | NT-3 growth factor receptor | Sus scrofa (Pig) | SS |
| P35739 | Ntrk1 | High affinity nerve growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q03351 | Ntrk3 | NT-3 growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q63604 | Ntrk2 | BDNF/NT-3 growth factors receptor | Rattus norvegicus (Rat) | SS |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGG3 | At1g51820 | Probable LRR receptor-like serine/threonine-protein kinase At1g51820 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGW2 | PAM74 | Probable LRR receptor-like serine/threonine-protein kinase PAM74 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FN93 | At5g59680 | Probable LRR receptor-like serine/threonine-protein kinase At5g59680 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FN94 | At5g59670 | Receptor-like protein kinase At5g59670 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LIG2 | At3g21340 | Receptor-like protein kinase At3g21340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZQR3 | At2g14510 | Leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14510 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LPZ9 | SD113 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P43298 | TMK1 | Receptor protein kinase TMK1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGI2 | At1g67720 | Probable LRR receptor-like serine/threonine-protein kinase At1g67720 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2S4 | LECRKS4 | L-type lectin-domain containing receptor kinase S.4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWZ5 | SD25 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64556 | At2g19230 | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g19230 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SXB4 | At1g11300 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q84JQ4 | PRK2 | Pollen receptor-like kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q93ZS4 | NIK3 | Protein NSP-INTERACTING KINASE 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94AG2 | SERK1 | Somatic embryogenesis receptor kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64477 | At2g19130 | G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9XIC7 | SERK2 | Somatic embryogenesis receptor kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEFPHSVLLV | VLIIATFAIS | NLVQAEEDQE | GFISLDCGLP | PNEVSPYIEP | FTGLRFSSDS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SFIQSGKIGK | VDKSFEATTL | KSYMTLRYFP | DGKRNCYNLI | VKQGKTYMIR | ATALYGNYDG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LNISPKFDLY | IGANFWTTLD | AGEYLSGVVE | EVNYIPRSNS | LDVCLVKTDT | STPFLSLLEL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RPLDNDSYLT | GSGSLKTFRR | YYLSNSESVI | AYPEDVKDRI | WEPTFDSEWK | QIWTTLKPNN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| SNGYLVPKNV | LMTAAIPAND | SAPFRFTEEL | DSPTDELYVY | LHFSEVQSLQ | ANESREFDIL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| WSGEVAYEAF | IPEYLNITTI | QTNTPVTCPG | GKCNLELKRT | KNSTHPPLIN | AIEFYTVVNF |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PQLETNETDV | VAIKDIKATY | ELNRITWQGD | PCVPQKFIWE | GLDCNSKDAL | TLPRITSLNL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SSTGLTGNIA | AGIQNLTHLD | KLDLSNNNLT | GGVPEFLASM | KSLSFINLSK | NNLNGSIPQA |
| 490 | 500 | 510 | 520 | 530 | 540 |
| LLKREKDGLK | LSVDEQIRCF | PGSCVITKKK | FPVMIVALVS | SAVVVILVVL | VLIFVFKKKK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| PSNLEDLPPS | SNTPRENITS | TSISDTSIET | KRKRFSYSEV | MEMTKNLQRP | LGEGGFGVVY |
| 610 | 620 | 630 | 640 | 650 | 660 |
| HGDINGSSQQ | VAVKLLSQSS | TQGYKEFKAE | VELLLRVHHI | NLVSLVGYCD | ERDHLALIYE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| YMSNKDLKHH | LSGKHGGSVL | KWNTRLQIAV | DAALGLEYLH | IGCRPSMVHR | DVKSTNILLD |
| 730 | 740 | 750 | 760 | 770 | 780 |
| DQFTAKMADF | GLSRSFQLGD | ESQVSTVVAG | TPGYLDPEYY | RTGRLAEMSD | VYSFGIVLLE |
| 790 | 800 | 810 | 820 | 830 | 840 |
| IITNQRVIDP | AREKSHITEW | TAFMLNRGDI | TRIMDPNLQG | DYNSRSVWRA | LELAMMCANP |
| 850 | 860 | 870 | 880 | ||
| SSEKRPSMSQ | VVIELKECIR | SENKTQGMDS | HSSFEQSMSF | DTKAVPSAR |