Q9SHT3
Gene name |
At2g05410 (F16J10.4) |
Protein name |
MATH domain and coiled-coil domain-containing protein At2g05410 |
Names |
RTM3-like protein At2g05410 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT2G05410 |
EC number |
|
Protein Class |
|
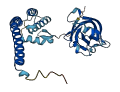
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9SHT3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9SHT3-F1 | Predicted | AlphaFoldDB |
88 variants for Q9SHT3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH01653997 | 4 | T>I | No | 1000Genomes | |
| ENSVATH11291376 | 7 | T>M | No | 1000Genomes | |
| tmp_2_1977241_T_C | 13 | I>T | No | 1000Genomes | |
| ENSVATH01653998 | 15 | N>I | No | 1000Genomes | |
| ENSVATH11291377 | 16 | F>L | No | 1000Genomes | |
| ENSVATH01653999 | 17 | S>Y | No | 1000Genomes | |
| ENSVATH05265543 | 19 | L>F | No | 1000Genomes | |
| tmp_2_1977265_C_G | 21 | S>C | No | 1000Genomes | |
| tmp_2_1977268_C_A | 22 | A>E | No | 1000Genomes | |
| ENSVATH01654000 | 23 | P>L | No | 1000Genomes | |
| ENSVATH00209042 | 23 | P>T | No | 1000Genomes | |
| ENSVATH11291378 | 25 | Y>C | No | 1000Genomes | |
| tmp_2_1977280_C_T | 26 | S>F | No | 1000Genomes | |
| ENSVATH05265545 | 27 | D>G | No | 1000Genomes | |
| ENSVATH05265547 | 28 | I>R | No | 1000Genomes | |
| ENSVATH05265548 | 32 | G>C | No | 1000Genomes | |
| ENSVATH14830455 | 36 | W>C | No | 1000Genomes | |
| tmp_2_1977310_G_T | 36 | W>L | No | 1000Genomes | |
| ENSVATH05265564 | 42 | P>L | No | 1000Genomes | |
| ENSVATH05265566 | 48 | A>G | No | 1000Genomes | |
| ENSVATH05265565 | 48 | A>T | No | 1000Genomes | |
| tmp_2_1977533_C_T | 53 | S>F | No | 1000Genomes | |
| ENSVATH01654030 | 58 | V>I | No | 1000Genomes | |
| ENSVATH13999860 | 59 | P>L | No | 1000Genomes | |
| ENSVATH01654031 | 66 | T>S | No | 1000Genomes | |
| tmp_2_1977574_G_C | 67 | G>R | No | 1000Genomes | |
| ENSVATH01654032 | 69 | R>Q | No | 1000Genomes | |
| ENSVATH00209047 | 73 | K>R | No | 1000Genomes | |
| ENSVATH05265569 | 74 | V>A | No | 1000Genomes | |
| tmp_2_1977623_C_T | 83 | S>L | No | 1000Genomes | |
| tmp_2_1977625_G_A | 84 | E>K | No | 1000Genomes | |
| tmp_2_1977630_A_C | 85 | K>N | No | 1000Genomes | |
| ENSVATH05265570 | 89 | R>* | No | 1000Genomes | |
| ENSVATH11291527 | 93 | Q>H | No | 1000Genomes | |
| tmp_2_1977771_G_C | 97 | V>L | No | 1000Genomes | |
| tmp_2_1977781_C_A | 100 | A>D | No | 1000Genomes | |
| tmp_2_1977789_TG_GG,T | 103 | W>G | No | 1000Genomes | |
| ENSVATH01654035 | 106 | P>R | No | 1000Genomes | |
| tmp_2_1977813_C_G | 111 | H>D | No | 1000Genomes | |
| tmp_2_1977823_T_C | 114 | L>P | No | 1000Genomes | |
| ENSVATH00209048 | 118 | M>K | No | 1000Genomes | |
| tmp_2_1977858_C_G | 126 | L>V | No | 1000Genomes | |
| ENSVATH01654037 | 129 | V>A | No | 1000Genomes | |
| ENSVATH01654038 | 145 | K>E | No | 1000Genomes | |
| tmp_2_1977925_C_A | 148 | S>* | No | 1000Genomes | |
| ENSVATH11291532 | 150 | I>R | No | 1000Genomes | |
| ENSVATH13999862 | 154 | I>V | No | 1000Genomes | |
| ENSVATH11291534 | 155 | D>E | No | 1000Genomes | |
| tmp_2_1977958_T_A | 159 | F>Y | No | 1000Genomes | |
| tmp_2_1978144_C_T | 168 | S>F | No | 1000Genomes | |
| ENSVATH01654045 | 168 | S>T | No | 1000Genomes | |
| ENSVATH01654046 | 185 | L>V | No | 1000Genomes | |
| ENSVATH05265577 | 186 | K>I | No | 1000Genomes | |
| ENSVATH08761422 | 187 | N>D | No | 1000Genomes | |
| tmp_2_1978209_T_A | 190 | L>M | No | 1000Genomes | |
| tmp_2_1978230_C_T | 197 | L>F | No | 1000Genomes | |
| ENSVATH05265579 | 200 | S>T | No | 1000Genomes | |
| tmp_2_1978255_T_A | 205 | L>Q | No | 1000Genomes | |
| tmp_2_1978258_G_T | 206 | C>F | No | 1000Genomes | |
| ENSVATH11291816 | 208 | S>L | No | 1000Genomes | |
| ENSVATH05265580 | 211 | E>G | No | 1000Genomes | |
| ENSVATH00209052 | 219 | D>H | No | 1000Genomes | |
| ENSVATH01654051 | 220 | A>T | No | 1000Genomes | |
| ENSVATH13999946 | 221 | G>D | No | 1000Genomes | |
| tmp_2_1978309_C_A | 223 | A>E | No | 1000Genomes | |
| ENSVATH01654052 | 234 | L>M | No | 1000Genomes | |
| tmp_2_1978344_G_T | 235 | D>Y | No | 1000Genomes | |
| ENSVATH01654053 | 236 | W>* | No | 1000Genomes | |
| ENSVATH05265583 | 239 | K>N | No | 1000Genomes | |
| tmp_2_1978368_G_A | 243 | E>K | No | 1000Genomes | |
| tmp_2_1978376_G_C | 245 | K>N | No | 1000Genomes | |
| ENSVATH01654054 | 246 | E>G | No | 1000Genomes | |
| tmp_2_1978385_G_C | 248 | K>N | No | 1000Genomes | |
| ENSVATH11291817 | 251 | E>* | No | 1000Genomes | |
| ENSVATH11291817 | 251 | E>K | No | 1000Genomes | |
| ENSVATH01654055 | 253 | S>A | No | 1000Genomes | |
| ENSVATH01654056 | 256 | V>A | No | 1000Genomes | |
| tmp_2_1978434_C_A | 265 | L>I | No | 1000Genomes | |
| ENSVATH11291818 | 266 | Q>* | No | 1000Genomes | |
| tmp_2_1978447_A_G | 269 | K>R | No | 1000Genomes | |
| ENSVATH01654062 | 281 | K>T | No | 1000Genomes | |
| ENSVATH13999947 | 282 | E>K | No | 1000Genomes | |
| ENSVATH01654063 | 286 | L>F | No | 1000Genomes | |
| tmp_2_1978497_T_A | 286 | L>M | No | 1000Genomes | |
| ENSVATH05265585 | 288 | A>V | No | 1000Genomes | |
| ENSVATH05265586 | 296 | Y>* | No | 1000Genomes | |
| ENSVATH05265587 | 299 | I>N | No | 1000Genomes | |
| ENSVATH13999948 | 301 | N>S | No | 1000Genomes |
No associated diseases with Q9SHT3
1 regional properties for Q9SHT3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | MATH/TRAF domain | 6 - 133 | IPR002083 |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9ZUA7 | At2g01790 | MATH domain and coiled-coil domain-containing protein At2g01790 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2J0 | At3g58260 | MATH domain and coiled-coil domain-containing protein At3g58260 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSD2 | At3g27040 | MATH domain and coiled-coil domain-containing protein At3g27040 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4IN32 | At2g42460 | MATH domain and coiled-coil domain-containing protein At2g42460 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0DKG4 | At2g42465 | MATH domain and coiled-coil domain-containing protein At2g42465 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0DKG7 | At2g42480 | MATH domain and coiled-coil domain-containing protein At2g42480 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4J4A0 | At3g44790 | MATH domain and coiled-coil domain-containing protein At3g44790 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4J4A1 | At3g44800 | MATH domain and coiled-coil domain-containing protein At3g44800 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2J5 | At3g58210 | MATH domain and coiled-coil domain-containing protein At3g58210 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2I2 | At3g58340 | MATH domain and coiled-coil domain-containing protein At3g58340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4J4P8 | At3g58220 | MATH domain and coiled-coil domain-containing protein At3g58220 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| METTVDTTIT | WVIKNFSSLQ | SAPIYSDIFV | VGGCKWRIMA | YPKGINKAHD | SFSLFLNVPD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NESLPTGWRR | HAKVSFSLVN | QGSEKLSQRK | VTQHWFVQKA | FTWGFPVMIT | HTELNAKMGF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LVNGELKVVA | KIEVLEVVGK | LDVSKESSPI | MKTIDVNGFQ | VLPSQVDSVK | RLFEKNLDIV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SKFRLKNPYL | KTACMNLLLS | LTETLCQSPQ | ELSNDGLSDA | GVALAYLIET | GLKLDWLEKK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LDELKEKKKK | EESCLVRLRE | MDEQLQPFKK | RCLDIEDQIS | KEKEELLAAR | EPLSLYDDID |
| NNV |