Q9S9M2
Gene name |
WAKL4 (At1g16150, T24D18.23) |
Protein name |
Wall-associated receptor kinase-like 4 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G16150 |
EC number |
|
Protein Class |
|
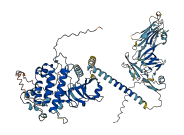
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
566-590 (Activation loop from InterPro)
Target domain |
424-703 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9S9M2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9S9M2-F1 | Predicted | AlphaFoldDB |
69 variants for Q9S9M2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH01057669 | 9 | Q>L | No | 1000Genomes | |
| ENSVATH04584556 | 14 | V>A | No | 1000Genomes | |
| ENSVATH04584557 | 23 | V>I | No | 1000Genomes | |
| ENSVATH01057671 | 31 | Y>D | No | 1000Genomes | |
| tmp_1_5532518_G_C | 35 | R>T | No | 1000Genomes | |
| tmp_1_5532520_G_T | 36 | V>F | No | 1000Genomes | |
| ENSVATH11314025 | 38 | G>R | No | 1000Genomes | |
| tmp_1_5532554_G_T | 47 | G>V | No | 1000Genomes | |
| tmp_1_5532563_G_A | 50 | G>E | No | 1000Genomes | |
| tmp_1_5532574_T_A | 54 | Y>N | No | 1000Genomes | |
| tmp_1_5532652_G_A | 80 | V>M | No | 1000Genomes | |
| ENSVATH13905007 | 97 | H>Q | No | 1000Genomes | |
| ENSVATH11314026 | 115 | L>R | No | 1000Genomes | |
| ENSVATH11314027 | 121 | N>S | No | 1000Genomes | |
| ENSVATH01057673 | 135 | N>S | No | 1000Genomes | |
| ENSVATH11314028 | 137 | L>F | No | 1000Genomes | |
| ENSVATH04584562 | 144 | V>A | No | 1000Genomes | |
| ENSVATH01057674 | 149 | M>T | No | 1000Genomes | |
| ENSVATH04584563 | 160 | S>* | No | 1000Genomes | |
| ENSVATH04584563 | 160 | S>L | No | 1000Genomes | |
| tmp_1_5532901_G_A | 163 | D>N | No | 1000Genomes | |
| ENSVATH01057675 | 172 | V>I | No | 1000Genomes | |
| tmp_1_5533157_A_G | 248 | D>G | No | 1000Genomes | |
| tmp_1_5533165_G_A | 251 | D>N | No | 1000Genomes | |
| ENSVATH00026252 | 253 | R>P | No | 1000Genomes | |
| ENSVATH04584566 | 275 | E>D | No | 1000Genomes | |
| tmp_1_5533258_T_G | 282 | Y>D | No | 1000Genomes | |
| tmp_1_5533292_G_A | 293 | C>Y | No | 1000Genomes | |
| ENSVATH04584567 | 312 | D>N | No | 1000Genomes | |
| tmp_1_5533525_G_T | 342 | K>N | No | 1000Genomes | |
| ENSVATH04584570 | 343 | K>Q | No | 1000Genomes | |
| ENSVATH04584571 | 346 | Q>K | No | 1000Genomes | |
| ENSVATH04584575 | 359 | A>S | No | 1000Genomes | |
| ENSVATH11314031 | 364 | A>S | No | 1000Genomes | |
| ENSVATH04584576 | 367 | I>N | No | 1000Genomes | |
| ENSVATH04584577 | 380 | S>I | No | 1000Genomes | |
| ENSVATH04584580 | 426 | T>K | No | 1000Genomes | |
| tmp_1_5533867_A_C | 426 | T>P | No | 1000Genomes | |
| ENSVATH11314033 | 435 | Q>K | No | 1000Genomes | |
| ENSVATH04584581 | 454 | S>* | No | 1000Genomes | |
| ENSVATH11314034 | 460 | D>Y | No | 1000Genomes | |
| ENSVATH01057677 | 461 | K>N | No | 1000Genomes | |
| ENSVATH04584582 | 464 | E>* | No | 1000Genomes | |
| tmp_1_5534041_T_A | 484 | L>M | No | 1000Genomes | |
| ENSVATH11314216 | 491 | E>D | No | 1000Genomes | |
| tmp_1_5534078_T_A | 496 | V>D | No | 1000Genomes | |
| ENSVATH00026254 | 513 | C>S | No | 1000Genomes | |
| ENSVATH13905009 | 538 | H>L | No | 1000Genomes | |
| ENSVATH04584586 | 568 | F>I | No | 1000Genomes | |
| ENSVATH00026256 | 577 | D>N | No | 1000Genomes | |
| ENSVATH11314218 | 582 | T>I | No | 1000Genomes | |
| ENSVATH13905010 | 613 | V>L | No | 1000Genomes | |
| ENSVATH04584590 | 615 | L>F | No | 1000Genomes | |
| ENSVATH04584592 | 623 | N>K | No | 1000Genomes | |
| tmp_1_5534521_G_A | 644 | V>M | No | 1000Genomes | |
| ENSVATH00026257 | 649 | F>V | No | 1000Genomes | |
| tmp_1_5534569_G_T | 660 | E>* | No | 1000Genomes | |
| ENSVATH11314220 | 661 | C>Y | No | 1000Genomes | |
| ENSVATH01057678 | 674 | K>E | No | 1000Genomes | |
| ENSVATH01057679 | 701 | S>P | No | 1000Genomes | |
| ENSVATH01057681 | 723 | A>S | No | 1000Genomes | |
| ENSVATH04584595 | 727 | N>K | No | 1000Genomes | |
| ENSVATH00026258 | 729 | E>D | No | 1000Genomes | |
| ENSVATH11314397 | 734 | V>I | No | 1000Genomes | |
| tmp_1_5534795_G_A | 735 | G>D | No | 1000Genomes | |
| ENSVATH13905013 | 738 | A>V | No | 1000Genomes | |
| tmp_1_5534809_G_C | 740 | A>P | No | 1000Genomes | |
| ENSVATH00026259 | 744 | N>S | No | 1000Genomes | |
| ENSVATH04584597 | 758 | L>Q | No | 1000Genomes |
No associated diseases with Q9S9M2
5 regional properties for Q9S9M2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 424 - 703 | IPR000719 |
| active_site | Serine/threonine-protein kinase, active site | 545 - 557 | IPR008271 |
| domain | Wall-associated receptor kinase | 174 - 282 | IPR013695 |
| conserved_site | EGF-like calcium-binding, conserved site | 312 - 339 | IPR018097 |
| domain | Wall-associated receptor kinase, galacturonan-binding domain | 33 - 74 | IPR025287 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| polysaccharide binding | Binding to a polysaccharide, a polymer of many (typically more than 10) monosaccharide residues linked glycosidically. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell surface receptor signaling pathway | The series of molecular signals initiated by activation of a receptor on the surface of a cell. The pathway begins with binding of an extracellular ligand to a cell surface receptor, or for receptors that signal in the absence of a ligand, by ligand-withdrawal or the activity of a constitutively active receptor. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
34 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P28693 | EPHB2 | Ephrin type-B receptor 2 | Gallus gallus (Chicken) | PR |
| P29318 | EPHA3 | Ephrin type-A receptor 3 | Gallus gallus (Chicken) | SS |
| Q07497 | EPHB5 | Ephrin type-B receptor 5 | Gallus gallus (Chicken) | PR |
| Q07496 | EPHA4 | Ephrin type-A receptor 4 | Gallus gallus (Chicken) | SS |
| O42422 | EPHA7 | Ephrin type-A receptor 7 | Gallus gallus (Chicken) | SS |
| Q07494 | EPHB1 | Ephrin type-B receptor 1 | Gallus gallus (Chicken) | SS |
| P54755 | EPHA5 | Ephrin type-A receptor 5 | Gallus gallus (Chicken) | SS |
| Q07498 | EPHB3 | Ephrin type-B receptor 3 | Gallus gallus (Chicken) | SS |
| P06213 | INSR | Insulin receptor | Homo sapiens (Human) | EV |
| P08069 | IGF1R | Insulin-like growth factor 1 receptor | Homo sapiens (Human) | EV |
| P14616 | INSRR | Insulin receptor-related protein | Homo sapiens (Human) | SS |
| P00533 | EGFR | Epidermal growth factor receptor | Homo sapiens (Human) | EV |
| Q15303 | ERBB4 | Receptor tyrosine-protein kinase erbB-4 | Homo sapiens (Human) | SS |
| P04626 | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | Homo sapiens (Human) | SS |
| P21860 | ERBB3 | Receptor tyrosine-protein kinase erbB-3 | Homo sapiens (Human) | SS |
| P05532 | Kit | Mast/stem cell growth factor receptor Kit | Mus musculus (Mouse) | PR |
| P26618 | Pdgfra | Platelet-derived growth factor receptor alpha | Mus musculus (Mouse) | SS |
| P35917 | Flt4 | Vascular endothelial growth factor receptor 3 | Mus musculus (Mouse) | SS |
| P35918 | Kdr | Vascular endothelial growth factor receptor 2 | Mus musculus (Mouse) | PR |
| P20786 | Pdgfra | Platelet-derived growth factor receptor alpha | Rattus norvegicus (Rat) | SS |
| Q05030 | Pdgfrb | Platelet-derived growth factor receptor beta | Rattus norvegicus (Rat) | SS |
| O08775 | Kdr | Vascular endothelial growth factor receptor 2 | Rattus norvegicus (Rat) | SS |
| Q498D6 | Fgfr4 | Fibroblast growth factor receptor 4 | Rattus norvegicus (Rat) | PR |
| Q04589 | Fgfr1 | Fibroblast growth factor receptor 1 | Rattus norvegicus (Rat) | SS |
| Q00495 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Rattus norvegicus (Rat) | PR |
| Q7X8C5 | WAKL2 | Wall-associated receptor kinase-like 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VYA3 | WAKL10 | Wall-associated receptor kinase-like 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C9L5 | WAKL9 | Wall-associated receptor kinase-like 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M092 | WAKL17 | Wall-associated receptor kinase-like 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LMN7 | WAK5 | Wall-associated receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9YI66 | tyro3 | Tyrosine-protein kinase receptor TYRO3 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKKETQNLQC | IPLVISVLSL | FGVSSARKPP | YLCNRVCGGI | SIPFPFGIGG | KECYLNPWYE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VVCNTTTSVP | FLSRINRELV | NIYLPDPTEY | YSNGVVHIKG | PVTSSGCSTG | TSQPLTPQPL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NVAGQGSPYF | LTDKNLLMAV | GCNVKAVMMD | VKSQIIGCES | SCDERNSSSQ | VVRNKICSGN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KCCQTRIPEG | QPQVIGVNIE | IPENKNTTEG | GCKVAFLTSN | KYSSLNVTEP | EEFHSDGYAV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VELGWYFDTS | DSRVLSPIGC | MNVSDASQDG | GYGSETICVC | SYGYFSGFSY | RSCYCNSMGY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| AGNPFLPGGC | VDIDECKLEI | GRKRCKDQSC | VNKPGWFTCE | PKKPGQIKPV | FQGVLIGSAL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LLFAFGIFGL | YKFIKKQRRS | SRMRVFFRRN | GGMLLKQQLA | RKEGNVEMSK | IFSSNELEKA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TDNFNTNRVL | GQGGQGTVYK | GMLVDGRIVA | VKRSKAMDED | KVEEFINEVV | VLAQINHRNI |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VKLLGCCLET | EVPVLVYEFV | PNGDLCKRLR | DECDDYIMTW | EVRLHIAIEI | AGALSYLHSA |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ASFPIYHRDI | KTTNILLDEK | YQVKVSDFGT | SRSVTIDQTH | LTTQVAGTFG | YVDPEYFQSS |
| 610 | 620 | 630 | 640 | 650 | 660 |
| KFTDKSDVYS | FGVVLVELIT | GKNPSSRVQS | EENRGFAAHF | VAAVKENRFL | DIVDERIKDE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| CNLDQVMAVA | KLAKRCLNRK | GKKRPNMREV | SVELERIRSS | SYKSEIHNDD | DDDDDDDDED |
| 730 | 740 | 750 | 760 | ||
| DQAMELNIEE | TWDVGMTAPA | SMFNNGSPAS | DVEPLVPLRT | W |