Q9QVP9
Gene name |
Ptk2b (Fak2, Pyk2, Raftk) |
Protein name |
Protein-tyrosine kinase 2-beta |
Names |
Calcium-dependent tyrosine kinase, CADTK, Calcium-regulated non-receptor proline-rich tyrosine kinase, Cell adhesion kinase beta, CAK-beta, CAKB, Focal adhesion kinase 2, FADK 2, Proline-rich tyrosine kinase 2, Related adhesion focal tyrosine kinase, RAFTK |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:19229 |
EC number |
2.7.10.2: Protein-tyrosine kinases |
Protein Class |
|
Descriptions
(Annotation based on sequence homology with Q00944)
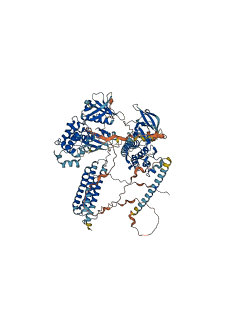
Focal adhesion kinase (FAK) integrates signals from integrin and growth factor receptors to regulate cellular responses including cell adhesion, migration and survival.
The inactive structure reveals a mechanism of inhibition in which the N-terminal FERM domain directly binds the kinase domain, blocking access to the catalytic cleft and protecting the FAK activation loop from Src phosphorylation. Additionally, the FERM domain sequesters the Tyr397 autophosphorylation and Src recruitment site, which lies in the linker connecting the FERM and kinase domains. FAK is activated by autophosphorylation at Tyr-397, which promotes interaction with Src and phosphorylation at Tyr-576 and Tyr-577 in the kinase activation loop.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
566-590 (Activation loop from InterPro)
Target domain |
425-683 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9QVP9
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9QVP9-F1 | Predicted | AlphaFoldDB |
47 variants for Q9QVP9
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389322302 | 7 | P>S | No | EVA | |
| rs3389344536 | 29 | V>L | No | EVA | |
| rs3389342045 | 91 | G>R | No | EVA | |
| rs3389335376 | 98 | K>E | No | EVA | |
| rs3389355470 | 125 | A>T | No | EVA | |
| rs1133696827 | 150 | T>R | No | EVA | |
| rs3389328653 | 157 | Q>P | No | EVA | |
| rs3389335378 | 164 | Q>* | No | EVA | |
| rs3389302925 | 171 | S>T | No | EVA | |
| rs3389292113 | 185 | R>I | No | EVA | |
| rs3389302973 | 200 | N>I | No | EVA | |
| rs3389309846 | 255 | A>S | No | EVA | |
| rs3389335303 | 268 | E>D | No | EVA | |
| rs3389302986 | 276 | T>I | No | EVA | |
| rs3389342026 | 297 | T>S | No | EVA | |
| rs3389344467 | 300 | A>D | No | EVA | |
| rs6378901 | 306 | K>R | No | EVA | |
| rs3389292190 | 331 | L>V | No | EVA | |
| rs3389351046 | 338 | L>V | No | EVA | |
| rs3389336992 | 341 | A>V | No | EVA | |
| rs3389355455 | 356 | G>V | No | EVA | |
| rs3389344515 | 364 | M>V | No | EVA | |
| rs3389340651 | 376 | L>M | No | EVA | |
| rs3389328674 | 382 | L>I | No | EVA | |
| rs3389340621 | 520 | S>Y | No | EVA | |
| rs3389302909 | 542 | S>C | No | EVA | |
| rs3389336947 | 632 | K>R | No | EVA | |
| rs3404979839 | 639 | E>* | No | EVA | |
| rs3389320144 | 641 | G>W | No | EVA | |
| rs3389339379 | 642 | D>N | No | EVA | |
| rs3389328713 | 662 | W>C | No | EVA | |
| rs3389340662 | 683 | Y>N | No | EVA | |
| rs3389335355 | 704 | I>R | No | EVA | |
| rs3405213146 | 776 | R>* | No | EVA | |
| rs3389309815 | 796 | L>F | No | EVA | |
| rs3389320146 | 798 | E>K | No | EVA | |
| rs3389340664 | 824 | R>Q | No | EVA | |
| rs3389292133 | 840 | P>S | No | EVA | |
| rs3389302955 | 845 | K>R | No | EVA | |
| rs3389262064 | 867 | I>V | No | EVA | |
| rs229138524 | 908 | V>A | No | EVA | |
| rs3389339386 | 910 | V>M | No | EVA | |
| rs252773007 | 926 | D>E | No | EVA | |
| rs3389309869 | 939 | I>T | No | EVA | |
| rs3389335328 | 952 | E>K | No | EVA | |
| rs213645523 | 958 | K>R | No | EVA | |
| rs3389339367 | 978 | T>I | No | EVA |
No associated diseases with Q9QVP9
12 regional properties for Q9QVP9
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | GPS motif | 686 - 738 | IPR000203 |
| domain | Cysteine-rich flanking region, C-terminal | 176 - 225 | IPR000483 |
| repeat | Leucine-rich repeat | 74 - 130 | IPR001611 |
| domain | GPCR, family 2, extracellular hormone receptor domain | 312 - 407 | IPR001879 |
| repeat | Leucine-rich repeat, typical subtype | 68 - 92 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 93 - 116 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 117 - 140 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 141 - 164 | IPR003591-4 |
| domain | Immunoglobulin subtype | 238 - 331 | IPR003599 |
| domain | Immunoglobulin-like domain | 231 - 329 | IPR007110 |
| domain | Immunoglobulin I-set | 236 - 329 | IPR013098 |
| domain | GPCR, family 2-like, transmembrane domain | 745 - 1039 | IPR017981 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.2 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
20 GO annotations of cellular component
| Name | Definition |
|---|---|
| apical dendrite | A dendrite that emerges near the apical pole of a neuron. In bipolar neurons, apical dendrites are located on the opposite side of the soma from the axon. |
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| cell body | The portion of a cell bearing surface projections such as axons, dendrites, cilia, or flagella that includes the nucleus, but excludes all cell projections. |
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| extrinsic component of cytoplasmic side of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to its cytoplasmic surface, but not integrated into the hydrophobic region. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| growth cone | The migrating motile tip of a growing neuron projection, where actin accumulates, and the actin cytoskeleton is the most dynamic. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| NMDA selective glutamate receptor complex | An assembly of four or five subunits which form a structure with an extracellular N-terminus and a large loop that together form the ligand binding domain. The C-terminus is intracellular. The ionotropic glutamate receptor complex itself acts as a ligand gated ion channel; on binding glutamate, charged ions pass through a channel in the center of the receptor complex. NMDA receptors are composed of assemblies of NR1 subunits (Figure 3) and NR2 subunits, which can be one of four separate gene products (NR2A-D). Expression of both subunits are required to form functional channels. The glutamate binding domain is formed at the junction of NR1 and NR2 subunits. NMDA receptors are permeable to calcium ions as well as being permeable to other ions. Thus NMDA receptor activation leads to a calcium influx into the post-synaptic cells, a signal thought to be crucial for the induction of NMDA-receptor dependent LTP and LTD. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| postsynapse | The part of a synapse that is part of the post-synaptic cell. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
13 GO annotations of molecular function
| Name | Definition |
|---|---|
| 3-phosphoinositide-dependent protein kinase binding | Binding to a 3-phosphoinositide-dependent protein kinase. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin-dependent protein kinase activity | Calmodulin-dependent catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; and ATP + a protein threonine = ADP + protein threonine phosphate. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| NMDA glutamate receptor activity | An cation channel that opens in response to binding by extracellular glutmate, but only if glycine is also bound and the membrane is depolarized. Voltage gating is indirect, due to ejection of bound magnesium from the pore at permissive voltages. |
| non-membrane spanning protein tyrosine kinase activity | Catalysis of the reaction: ATP + protein L-tyrosine = ADP + protein L-tyrosine phosphate by a non-membrane spanning protein. |
| protein C-terminus binding | Binding to a protein C-terminus, the end of a peptide chain at which the 1-carboxyl function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein self-association | Binding to a domain within the same polypeptide. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| protein-containing complex binding | Binding to a macromolecular complex. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
95 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| activation of GTPase activity | Any process that initiates the activity of an inactive GTPase through the replacement of GDP by GTP. |
| activation of Janus kinase activity | The process of introducing a phosphate group to a tyrosine residue of a JAK (Janus Activated Kinase) protein, thereby activating it. |
| adaptive immune response | An immune response mediated by cells expressing specific receptors for antigen produced through a somatic diversification process, and allowing for an enhanced secondary response to subsequent exposures to the same antigen (immunological memory). |
| angiogenesis | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels. |
| blood vessel endothelial cell migration | The orderly movement of an endothelial cell into the extracellular matrix in order to form new blood vessels during angiogenesis. |
| bone resorption | The process in which specialized cells known as osteoclasts degrade the organic and inorganic portions of bone, and endocytose and transport the degradation products. |
| cell adhesion | The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cell surface receptor signaling pathway | The series of molecular signals initiated by activation of a receptor on the surface of a cell. The pathway begins with binding of an extracellular ligand to a cell surface receptor, or for receptors that signal in the absence of a ligand, by ligand-withdrawal or the activity of a constitutively active receptor. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| cellular defense response | A defense response that is mediated by cells. |
| cellular response to fluid shear stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a fluid shear stress stimulus. Fluid shear stress is the force acting on an object in a system where the fluid is moving across a solid surface. |
| cellular response to retinoic acid | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a retinoic acid stimulus. |
| chemokine-mediated signaling pathway | The series of molecular signals initiated by a chemokine binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| endothelin receptor signaling pathway | A G protein-coupled receptor signaling pathway initiated by endothelin binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| epidermal growth factor receptor signaling pathway | The series of molecular signals initiated by binding of a ligand to the tyrosine kinase receptor EGFR (ERBB1) on the surface of a cell. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| focal adhesion assembly | The aggregation and bonding together of a set of components to form a focal adhesion, a complex of intracellular signaling and structural proteins that provides a structural link between the internal actin cytoskeleton and the ECM, and also function as a locus of signal transduction activity. |
| glial cell proliferation | The multiplication or reproduction of glial cells by cell division, resulting in the expansion of their population. Glial cells exist throughout the nervous system, and include Schwann cells, astrocytes, and oligodendrocytes among others. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| integrin-mediated signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to an integrin on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| ionotropic glutamate receptor signaling pathway | The series of molecular signals initiated by glutamate binding to a glutamate receptor on the surface of the target cell, followed by the movement of ions through a channel in the receptor complex, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| long-term synaptic depression | A process that modulates synaptic plasticity such that synapses are changed resulting in the decrease in the rate, or frequency of synaptic transmission at the synapse. |
| long-term synaptic potentiation | A process that modulates synaptic plasticity such that synapses are changed resulting in the increase in the rate, or frequency of synaptic transmission at the synapse. |
| MAPK cascade | An intracellular protein kinase cascade containing at least a MAPK, a MAPKK and a MAP3K. The cascade can also contain an additional tiers: the upstream MAP4K. The kinases in each tier phosphorylate and activate the kinase in the downstream tier to transmit a signal within a cell. |
| marginal zone B cell differentiation | The process in which a B cell in the spleen acquires the specialized features of a marginal zone B cell. Marginal zone B cells are localized in a distinct anatomical region of the spleen that represents the major antigen-filtering and scavenging area (by specialized macrophages resident there). It appears that they are preselected to express a BCR repertoire similar to B-1 B cells, biased toward bacterial cell wall constituents and senescent self-components (such as oxidized LDL). |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of bone mineralization | Any process that stops, prevents, or reduces the frequency, rate or extent of bone mineralization. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of muscle cell apoptotic process | Any process that decreases the rate or frequency of muscle cell apoptotic process, a form of programmed cell death induced by external or internal signals that trigger the activity of proteolytic caspases whose actions dismantle a muscle cell and result in its death. |
| negative regulation of myeloid cell differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of myeloid cell differentiation. |
| negative regulation of neuron apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process in neurons. |
| negative regulation of ossification | Any process that stops, prevents, or reduces the frequency, rate or extent of ossification, the formation of bone or of a bony substance or the conversion of fibrous tissue or of cartilage into bone or a bony substance. |
| negative regulation of potassium ion transport | Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of potassium ions (K+) into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| neuron projection development | The process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| oocyte maturation | A developmental process, independent of morphogenetic (shape) change, that is required for an oocyte to attain its fully functional state. Oocyte maturation commences after reinitiation of meiosis commonly starting with germinal vesicle breakdown, and continues up to the second meiotic arrest prior to fertilization. |
| peptidyl-tyrosine autophosphorylation | The phosphorylation by a protein of one or more of its own tyrosine amino acid residues, or a tyrosine residue on an identical protein. |
| peptidyl-tyrosine phosphorylation | The phosphorylation of peptidyl-tyrosine to form peptidyl-O4'-phospho-L-tyrosine. |
| positive regulation of actin filament polymerization | Any process that activates or increases the frequency, rate or extent of actin polymerization. |
| positive regulation of angiogenesis | Any process that activates or increases angiogenesis. |
| positive regulation of B cell chemotaxis | Any process that activates or increases the frequency, rate or extent of B cell chemotaxis. |
| positive regulation of cell growth | Any process that activates or increases the frequency, rate, extent or direction of cell growth. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of cell-matrix adhesion | Any process that activates or increases the rate or extent of cell adhesion to an extracellular matrix. |
| positive regulation of cytosolic calcium ion concentration | Any process that increases the concentration of calcium ions in the cytosol. |
| positive regulation of DNA biosynthetic process | Any process that activates or increases the frequency, rate or extent of DNA biosynthetic process. |
| positive regulation of endothelial cell migration | Any process that increases the rate, frequency, or extent of the orderly movement of an endothelial cell into the extracellular matrix to form an endothelium. |
| positive regulation of ERK1 and ERK2 cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| positive regulation of excitatory postsynaptic potential | Any process that enhances the establishment or increases the extent of the excitatory postsynaptic potential (EPSP) which is a temporary increase in postsynaptic potential due to the flow of positively charged ions into the postsynaptic cell. The flow of ions that causes an EPSP is an excitatory postsynaptic current (EPSC) and makes it easier for the neuron to fire an action potential. |
| positive regulation of JNK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the JNK cascade. |
| positive regulation of JUN kinase activity | Any process that activates or increases the frequency, rate or extent of JUN kinase activity. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| positive regulation of nitric oxide biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nitric oxide. |
| positive regulation of nitric-oxide synthase activity | Any process that activates or increases the activity of the enzyme nitric-oxide synthase. |
| positive regulation of peptidyl-tyrosine phosphorylation | Any process that activates or increases the frequency, rate or extent of the phosphorylation of peptidyl-tyrosine. |
| positive regulation of phosphatidylinositol 3-kinase activity | Any process that activates or increases the frequency, rate or extent of phosphatidylinositol 3-kinase activity. |
| positive regulation of protein kinase activity | Any process that activates or increases the frequency, rate or extent of protein kinase activity. |
| positive regulation of protein metabolic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein. |
| positive regulation of reactive oxygen species metabolic process | Any process that activates or increases the frequency, rate or extent of reactive oxygen species metabolic process. |
| positive regulation of synaptic transmission, glutamatergic | Any process that activates, maintains or increases the frequency, rate or extent of glutamatergic synaptic transmission, the process of communication from a neuron to another neuron across a synapse using the neurotransmitter glutamate. |
| positive regulation of translation | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| positive regulation of ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of ubiquitin-dependent protein catabolic process. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of actin cytoskeleton reorganization | Any process that modulates the frequency, rate or extent of actin cytoskeleton reorganization. |
| regulation of calcium-mediated signaling | Any process that modulates the frequency, rate or extent of calcium-mediated signaling, the process in which a cell uses calcium ions to convert an extracellular signal into a response. |
| regulation of cell adhesion | Any process that modulates the frequency, rate or extent of attachment of a cell to another cell or to the extracellular matrix. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of cGMP-mediated signaling | Any process that modulates the rate, frequency or extent of cGMP-mediated signaling. |
| regulation of establishment of cell polarity | Any process that modulates the frequency, rate or extent of establishment of cell polarity. |
| regulation of inositol trisphosphate biosynthetic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of inositol trisphosphate. |
| regulation of macrophage chemotaxis | Any process that modulates the rate, frequency or extent of macrophage chemotaxis. Macrophage chemotaxis is the movement of a macrophage in response to an external stimulus. |
| regulation of nitric oxide biosynthetic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nitric oxide. |
| regulation of NMDA receptor activity | Any process that modulates the frequency, rate or extent of N-methyl-D-aspartate selective glutamate receptor activity. |
| regulation of release of sequestered calcium ion into cytosol | Any process that modulates the frequency, rate or extent of the release into the cytosolic compartment of calcium ions sequestered in the endoplasmic reticulum or mitochondria. |
| regulation of ubiquitin-dependent protein catabolic process | Any process that modulates the frequency, rate or extent of ubiquitin-dependent protein catabolic process. |
| response to calcium ion | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a calcium ion stimulus. |
| response to cAMP | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate) stimulus. |
| response to cation stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of cation stress, an increase or decrease in the concentration of positively charged ions in the environment. |
| response to cocaine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cocaine stimulus. Cocaine is a crystalline alkaloid obtained from the leaves of the coca plant. |
| response to ethanol | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an ethanol stimulus. |
| response to glucose | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a glucose stimulus. |
| response to hormone | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hormone stimulus. |
| response to hydrogen peroxide | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hydrogen peroxide (H2O2) stimulus. |
| response to hypoxia | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension. Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%, results in metabolic adaptation at both the cellular and organismal level. |
| response to immobilization stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of being rendered immobile. |
| response to ischemia | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a inadequate blood supply. |
| response to lithium ion | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lithium (Li+) ion stimulus. |
| response to mechanical stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus. |
| response to xenobiotic stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a xenobiotic, a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
| signal complex assembly | The aggregation, arrangement and bonding together of a set of components to form a complex capable of relaying a signal within a cell. |
| sprouting angiogenesis | The extension of new blood vessels from existing vessels into avascular tissues, this process includes the specialization of endothelial cells into leading tip and stalk cells, proliferation and migration of the endothelial cells and cell adhesion resulting in angiogenic sprout fusion or lumen formation. |
| stress fiber assembly | The aggregation, arrangement and bonding together of a set of components to form a stress fiber. A stress fiber is a contractile actin filament bundle that consists of short actin filaments with alternating polarity. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| tumor necrosis factor-mediated signaling pathway | The series of molecular signals initiated by tumor necrosis factor binding to its receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| vascular endothelial growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a vascular endothelial growth factor receptor (VEGFR) on the surface of the target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
31 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q00944 | PTK2 | Focal adhesion kinase 1 | Gallus gallus (Chicken) | EV |
| Q05397 | PTK2 | Focal adhesion kinase 1 | Homo sapiens (Human) | EV |
| Q14289 | PTK2B | Protein-tyrosine kinase 2-beta | Homo sapiens (Human) | PR |
| P48025 | Syk | Tyrosine-protein kinase SYK | Mus musculus (Mouse) | SS |
| P43404 | Zap70 | Tyrosine-protein kinase ZAP-70 | Mus musculus (Mouse) | SS |
| Q64434 | Ptk6 | Protein-tyrosine kinase 6 | Mus musculus (Mouse) | SS |
| P14234 | Fgr | Tyrosine-protein kinase Fgr | Mus musculus (Mouse) | SS |
| P25911 | Lyn | Tyrosine-protein kinase Lyn | Mus musculus (Mouse) | EV |
| P39688 | Fyn | Tyrosine-protein kinase Fyn | Mus musculus (Mouse) | SS |
| P05480 | Src | Proto-oncogene tyrosine-protein kinase Src | Mus musculus (Mouse) | EV |
| P06240 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Mus musculus (Mouse) | SS |
| P16277 | Blk | Tyrosine-protein kinase Blk | Mus musculus (Mouse) | SS |
| P08103 | Hck | Tyrosine-protein kinase HCK | Mus musculus (Mouse) | SS |
| Q04736 | Yes1 | Tyrosine-protein kinase Yes | Mus musculus (Mouse) | SS |
| Q922K9 | Frk | Tyrosine-protein kinase FRK | Mus musculus (Mouse) | SS |
| Q03526 | Itk | Tyrosine-protein kinase ITK/TSK | Mus musculus (Mouse) | SS |
| P35991 | Btk | Tyrosine-protein kinase BTK | Mus musculus (Mouse) | EV |
| P24604 | Tec | Tyrosine-protein kinase Tec | Mus musculus (Mouse) | SS |
| P41242 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| P41241 | Csk | Tyrosine-protein kinase CSK | Mus musculus (Mouse) | EV |
| P34152 | Ptk2 | Focal adhesion kinase 1 | Mus musculus (Mouse) | SS |
| Q99ML2 | Tnk1 | Non-receptor tyrosine-protein kinase TNK1 | Mus musculus (Mouse) | PR |
| O54967 | Tnk2 | Activated CDC42 kinase 1 | Mus musculus (Mouse) | SS |
| P70451 | Fer | Tyrosine-protein kinase Fer | Mus musculus (Mouse) | PR |
| Q6J9G1 | Styk1 | Tyrosine-protein kinase STYK1 | Mus musculus (Mouse) | PR |
| P00520 | Abl1 | Tyrosine-protein kinase ABL1 | Mus musculus (Mouse) | EV |
| Q62270 | Srms | Tyrosine-protein kinase Srms | Mus musculus (Mouse) | SS |
| Q4JIM5 | Abl2 | Tyrosine-protein kinase ABL2 | Mus musculus (Mouse) | SS |
| O35346 | Ptk2 | Focal adhesion kinase 1 | Rattus norvegicus (Rat) | SS |
| P70600 | Ptk2b | Protein-tyrosine kinase 2-beta | Rattus norvegicus (Rat) | PR |
| Q95YD4 | kin-32 | Inactive tyrosine-protein kinase kin-32 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSGVSEPLSR | VKVGTLRRPE | GPPEPMVVVP | VDVEKEDVRI | LKVCFYSNSF | NPGKNFKLVK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CTVQTEIQEI | ITSILLSGRI | GPNIQLAECY | GLRLKHMKSD | EIHWLHPQMT | VGEVQDKYEC |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LHVEAEWRYD | LQIRYLPEDF | MESLKEDRTT | LLYFYQQLRN | DYMQRYASKV | SEGMALQLGC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LELRRFFKDM | PHNALDKKSN | FELLEKEVGL | DLFFPKQMQE | NLKPKQFRKM | IQQTFQQYAS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LREEECVMKF | FNTLAGFANI | DQETYRCELI | QGWNITVDLV | IGPKGIRQLT | SQDTKPTCLA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EFKQIKSIRC | LPLEETQAVL | QLGIEGAPQS | LSIKTSSLAE | AENMADLIDG | YCRLQGEHKG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SLIMHAKKDG | EKRNSLPQIP | TLNLEARRSH | LSESCSIESD | IYAEIPDETL | RRPGGPQYGV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AREEVVLNRI | LGEGFFGEVY | EGVYTNHKGE | KINVAVKTCK | KDCTQDNKEK | FMSEAVIMKN |
| 490 | 500 | 510 | 520 | 530 | 540 |
| LDHPHIVKLI | GIIEEEPTWI | IMELYPYGEL | GHYLERNKNS | LKVPTLVLYT | LQICKAMAYL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ESINCVHRDI | AVRNILVASP | ECVKLGDFGL | SRYIEDEDYY | KASVTRLPIK | WMSPESINFR |
| 610 | 620 | 630 | 640 | 650 | 660 |
| RFTTASDVWM | FAVCMWEILS | FGKQPFFWLE | NKDVIGVLEK | GDRLPKPELC | PPVLYTLMTR |
| 670 | 680 | 690 | 700 | 710 | 720 |
| CWDYDPSDRP | RFTELVCSLS | DIYQMEKDIA | IEQERNARYR | PPKILEPTTF | QEPPPKPSRP |
| 730 | 740 | 750 | 760 | 770 | 780 |
| KYRPPPQTNL | LAPKLQFQVP | EGLCASSPTL | TSPMEYPSPV | NSLHTPPLHR | HNVFKRHSMR |
| 790 | 800 | 810 | 820 | 830 | 840 |
| EEDFIRPSSR | EEAQQLWEAE | KIKMKQVLER | QQKQMVEDSQ | WLRREERCLD | PMVYMNDKSP |
| 850 | 860 | 870 | 880 | 890 | 900 |
| LTPEKEAGYT | EFTGPPQKPP | RLGAQSIQPT | ANLDRTDDLV | YHNVMTLVEA | VLELKNKLGQ |
| 910 | 920 | 930 | 940 | 950 | 960 |
| LPPEDYVVVV | KNVGLNLRKL | IGSVDDLLPS | LPASSRTEIE | GTQKLLNKDL | AELINKMKLA |
| 970 | 980 | 990 | 1000 | ||
| QQNAVTSLSE | DCKRQMLTAS | HTLAVDAKNL | LDAVDQAKVV | ANLAHPPAE |