Q9NRH2
Gene name |
SNRK (KIAA0096, SNFRK) |
Protein name |
SNF-related serine/threonine-protein kinase |
Names |
EC 2.7.11.1 , SNF1-related kinase |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:54861 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
16-269 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
157-179 (Activation loop from InterPro)
Target domain |
16-269 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Pang T et al. (2007) "Conserved alpha-helix acts as autoinhibitory sequence in AMP-activated protein kinase alpha subunits", The Journal of biological chemistry, 282, 495-506
- Hardie DG et al. (2012) "AMP-activated protein kinase: a target for drugs both ancient and modern", Chemistry & biology, 19, 1222-36
- Li YY et al. (2013) "Novel small-molecule AMPK activator orally exerts beneficial effects on diabetic db/db mice", Toxicology and applied pharmacology, 273, 325-34
- Oakhill JS et al. (2009) "Structure and function of AMP-activated protein kinase", Acta physiologica (Oxford, England), 196, 3-14
- Pang T et al. (2008) "Small molecule antagonizes autoinhibition and activates AMP-activated protein kinase in cells", The Journal of biological chemistry, 283, 16051-60
- Scott JW et al. (2014) "ATP sensitive bi-quinoline activator of the AMP-activated protein kinase", Biochemical and biophysical research communications, 443, 435-40
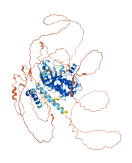
Autoinhibited structure

Activated structure

2 structures for Q9NRH2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 5YKS | X-ray | 290 A | A/B | 1-356 | PDB |
| AF-Q9NRH2-F1 | Predicted | AlphaFoldDB |
578 variants for Q9NRH2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| TCGA novel | 3 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1324272178 | 3 | G>E | No | TOPMed | |
| rs1249430769 | 4 | F>C | No | gnomAD | |
| rs1282382145 | 4 | F>V | No | TOPMed | |
| rs2090911554 | 5 | K>M | No | Ensembl | |
| rs754086109 | 7 | G>V | No |
ExAC gnomAD |
|
| rs1559460498 | 8 | Y>C | No | Ensembl | |
| rs2090911680 | 10 | G>R | No | TOPMed | |
| TCGA novel | 11 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 12 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2090911707 | 12 | I>M | No | gnomAD | |
| TCGA novel | 17 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1462617232 | 23 | G>S | No | gnomAD | |
| rs139338365 | 24 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs780505511 | 26 | H>R | No |
ExAC gnomAD |
|
| rs1429357100 | 27 | F>S | No |
TOPMed gnomAD |
|
|
COSM106817 rs143570625 |
28 | A>V | skin [Cosmic] | No |
cosmic curated Ensembl |
| rs754467995 | 29 | V>M | No |
ExAC TOPMed gnomAD |
|
| COSM5131700 | 30 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2125620201 | 30 | V>F | No | Ensembl | |
| TCGA novel | 33 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs2090912197 COSM1226998 COSM1226997 |
38 | T>M | large_intestine [Cosmic] | No |
cosmic curated Ensembl |
| rs2090912306 | 41 | K>N | No | Ensembl | |
| rs747774782 | 47 | I>V | No |
ExAC gnomAD |
|
| rs749271551 | 54 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs749271551 | 54 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1429171638 | 55 | L>Q | No | gnomAD | |
| rs373410633 | 55 | L>V | No |
ESP ExAC gnomAD |
|
| TCGA novel | 56 | A>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2090912570 | 58 | G>D | No | Ensembl | |
| rs1371436206 | 59 | H>Y | No | gnomAD | |
| rs1467713865 | 61 | F>S | No | gnomAD | |
|
COSM4911673 COSM4911672 |
67 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1188403194 | 67 | M>L | No |
TOPMed gnomAD |
|
| rs774409595 | 67 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1406526593 | 69 | L>P | No |
TOPMed gnomAD |
|
| rs1363931438 | 73 | P>S | No |
TOPMed gnomAD |
|
| rs2090912866 | 74 | N>S | No | TOPMed | |
| rs764166971 | 75 | I>V | No |
ExAC gnomAD |
|
| rs2090912958 | 76 | V>I | No | TOPMed | |
| rs1305274728 | 77 | R>C | No | gnomAD | |
| rs1224957129 | 77 | R>H | No | gnomAD | |
| rs1244214662 | 86 | T>I | No | gnomAD | |
| rs770438152 | 88 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1259673589 | 90 | L>I | No | gnomAD | |
| TCGA novel | 97 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 99 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1428917743 | 105 | M>V | No | gnomAD | |
| rs569432019 | 109 | E>V | No |
1000Genomes TOPMed |
|
| rs1173649669 | 112 | N>S | No | gnomAD | |
| rs989826664 | 114 | D>H | No | gnomAD | |
| rs779596849 | 115 | L>M | No |
TOPMed gnomAD |
|
| rs2090913662 | 119 | Y>C | No |
TOPMed gnomAD |
|
| TCGA novel | 121 | A>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1359253380 | 126 | A>G | No | gnomAD | |
| rs369827087 | 133 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1349439464 | 134 | H>R | No | gnomAD | |
|
COSM3594153 COSM3594152 |
134 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752317792 | 136 | V>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 138 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 143 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM5120799 | 144 | N>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1559460681 | 145 | V>A | No | Ensembl | |
| rs1259553236 | 147 | F>V | No | gnomAD | |
| rs1559460688 | 150 | K>R | No | Ensembl | |
| rs2090914168 | 151 | Q>R | No | TOPMed | |
| rs1200176044 | 152 | G>D | No |
TOPMed gnomAD |
|
| rs2090914262 | 156 | L>V | No | TOPMed | |
| TCGA novel | 158 | D>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763845809 | 163 | N>H | No | Ensembl | |
|
COSM3427602 COSM3427603 |
164 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 168 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2090914518 | 169 | K>R | No |
TOPMed gnomAD |
|
| rs2090914542 | 170 | K>T | No | TOPMed | |
| rs970946153 | 172 | T>N | No | TOPMed | |
| rs1426495654 | 174 | S>I | No | gnomAD | |
|
COSM4117689 COSM4117688 |
175 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2090914853 | 182 | A>T | No | gnomAD | |
| rs1339934466 | 184 | E>G | No | Ensembl | |
| rs1233242718 | 185 | I>V | No | Ensembl | |
| rs2090914990 | 187 | L>I | No | Ensembl | |
| rs1305345678 | 189 | D>G | No |
TOPMed gnomAD |
|
| rs1291059796 | 190 | E>G | No |
TOPMed gnomAD |
|
|
COSM1423184 COSM1423185 |
194 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3373185 COSM3373186 |
196 | V>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752077383 | 196 | V>I | No |
ExAC gnomAD |
|
| rs2090915197 | 197 | D>H | No |
TOPMed gnomAD |
|
| rs1575554085 | 201 | L>R | No | Ensembl | |
|
COSM188650 rs1233445337 |
209 | V>A | large_intestine [Cosmic] | No |
cosmic curated Ensembl |
| rs1233445337 | 209 | V>G | No | Ensembl | |
| rs1575554102 | 210 | C>W | No | Ensembl | |
| rs753133251 | 213 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs75494032 | 216 | Q>K | No | Ensembl | |
| TCGA novel | 217 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs539932047 | 218 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs2091151810 | 218 | A>V | No | TOPMed | |
| rs559568271 | 219 | N>S | No |
1000Genomes ExAC gnomAD |
|
| rs2091151882 | 221 | S>G | No | TOPMed | |
| TCGA novel | 222 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs370490912 | 226 | M>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 230 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756930855 | 231 | K>R | No |
ExAC gnomAD |
|
| rs2125639287 | 235 | P>A | No | Ensembl | |
| rs571878326 | 236 | S>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1272593106 | 237 | H>R | No | TOPMed | |
| rs780104262 | 240 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs780104262 | 240 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs1338610214 | 241 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1325377834 | 243 | K>R | No |
TOPMed gnomAD |
|
| rs1308982667 | 244 | D>G | No | TOPMed | |
| TCGA novel | 246 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761384780 | 248 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1284915054 | 248 | R>W | No |
TOPMed gnomAD |
|
| rs777706778 | 249 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs532190761 | 252 | R>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs368466705 | 253 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM263797 COSM263798 |
253 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2091225648 | 255 | K>* | No | TOPMed | |
| rs1471400535 | 256 | R>K | No | gnomAD | |
| rs1575559567 | 258 | A>T | No | TOPMed | |
|
rs35624204 VAR_041096 |
260 | L>S | No |
UniProt ExAC TOPMed dbSNP gnomAD |
|
| rs2091225766 | 260 | L>V | No | Ensembl | |
| rs1418434343 | 262 | E>K | No | gnomAD | |
|
COSM2157230 COSM2157229 |
263 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs955128936 | 264 | E>G | No | TOPMed | |
| rs2091225916 | 265 | N>K | No | TOPMed | |
| rs1366287768 | 266 | H>R | No | gnomAD | |
|
COSM3594159 COSM3594158 |
267 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1220249396 | 273 | D>N | No |
TOPMed gnomAD |
|
| rs372084850 | 280 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1289779656 | 281 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
COSM1044843 COSM1044841 |
281 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779174593 | 281 | N>T | No |
ExAC gnomAD |
|
| rs1344936905 | 282 | I>F | No | TOPMed | |
| TCGA novel | 283 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3594160 COSM3594161 |
286 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2125645511 | 287 | Y>H | No | Ensembl | |
| rs866480307 | 288 | K>R | No | Ensembl | |
| TCGA novel | 288 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091226279 | 289 | N>S | No | Ensembl | |
| TCGA novel | 289 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772506538 | 290 | L>F | No |
ExAC gnomAD |
|
|
COSM4926100 COSM4926099 |
290 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs569406710 | 291 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs879228122 | 294 | E>Q | No | Ensembl | |
| rs2091226448 | 297 | S>G | No | Ensembl | |
| TCGA novel | 297 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091226472 | 298 | I>L | No | TOPMed | |
| rs2091226494 | 299 | I>F | No | TOPMed | |
| rs769334631 | 301 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs552460740 | 301 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1313576307 | 303 | V>L | No |
TOPMed gnomAD |
|
| rs2091226667 | 308 | A>V | No | TOPMed | |
|
COSM1423186 rs889935483 COSM1423187 |
310 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs917888418 | 312 | A>S | No |
TOPMed gnomAD |
|
| rs917888418 | 312 | A>T | No |
TOPMed gnomAD |
|
| TCGA novel | 312 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766230368 | 314 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs2091251638 | 316 | A>V | No | Ensembl | |
| rs1559471313 | 317 | L>M | No | Ensembl | |
| rs1444359151 | 320 | N>S | No | gnomAD | |
| rs762497503 | 321 | R>K | No | ExAC | |
| rs1338600221 | 321 | R>S | No | gnomAD | |
| rs2091251744 | 322 | Y>C | No | Ensembl | |
| rs78507885 | 326 | T>P | No | Ensembl | |
|
COSM3427604 COSM3427605 rs2091251806 |
327 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| TCGA novel | 328 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2091251829 | 328 | T>I | No | TOPMed | |
|
COSM446523 COSM446524 |
334 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767234882 | 335 | R>T | No |
ExAC gnomAD |
|
| rs1227697345 | 336 | I>F | No | gnomAD | |
| rs1227697345 | 336 | I>L | No | gnomAD | |
| rs1257075760 | 336 | I>T | No | gnomAD | |
| TCGA novel | 337 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1452872989 | 338 | R>K | No | gnomAD | |
| rs2091252175 | 341 | Q>P | No | TOPMed | |
| rs1363283384 | 342 | E>Q | No |
TOPMed gnomAD |
|
| TCGA novel | 344 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1404416885 | 344 | E>D | No | gnomAD | |
| rs2091252258 | 345 | I>K | No | TOPMed | |
| rs1264175070 | 345 | I>M | No |
TOPMed gnomAD |
|
| rs760563902 | 346 | Q>K | No |
ExAC gnomAD |
|
| rs750516712 | 347 | T>N | No |
ExAC gnomAD |
|
| rs71323192 | 347 | T>P | No | Ensembl | |
| rs199515297 | 348 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 348 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751761706 | 351 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs755404685 | 352 | P>L | No |
ExAC gnomAD |
|
| TCGA novel | 353 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1559471382 | 354 | N>D | No | Ensembl | |
| rs748698874 | 354 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1559471385 | 355 | I>F | No | Ensembl | |
| rs1559471385 | 355 | I>L | No | Ensembl | |
| TCGA novel | 357 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs917563757 | 357 | A>V | No | Ensembl | |
| rs778347997 | 365 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1490877913 | 365 | T>N | No | gnomAD | |
| rs778347997 | 365 | T>S | No |
ExAC TOPMed gnomAD |
|
|
COSM4413973 COSM4413974 |
366 | K>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1472217782 | 366 | K>R | No |
TOPMed gnomAD |
|
| rs2091284411 | 368 | D>H | No | Ensembl | |
| rs771759736 | 370 | P>S | No |
ExAC gnomAD |
|
| rs2125650815 | 372 | D>A | No | Ensembl | |
| TCGA novel | 373 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1575564117 | 376 | D>A | No | Ensembl | |
|
COSM1485194 COSM1485193 |
376 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs377372127 | 378 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4117693 rs377372127 COSM4117692 |
378 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs377372127 | 378 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1575564131 | 380 | T>I | No | Ensembl | |
|
COSM4849026 COSM4849027 |
381 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs369083124 | 381 | P>R | No | ESP | |
| TCGA novel | 382 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 383 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776443103 | 385 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs776443103 | 385 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs183015402 | 385 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs774616197 | 386 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs774616197 | 386 | T>S | No |
ExAC TOPMed gnomAD |
|
| COSM72749 | 387 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs56104180 VAR_041097 |
391 | P>S | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1240041340 | 392 | A>T | No | gnomAD | |
| rs1307766065 | 392 | A>V | No | gnomAD | |
| rs752967293 | 393 | R>Q | No |
ExAC gnomAD |
|
|
COSM347262 COSM347261 rs370283116 |
393 | R>W | lung [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1289281753 | 396 | D>N | No | gnomAD | |
| rs2091285393 | 396 | D>V | No | Ensembl | |
| rs1320071461 | 397 | S>G | No |
TOPMed gnomAD |
|
| TCGA novel | 399 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756439526 | 400 | N>I | No |
ExAC gnomAD |
|
| rs756439526 | 400 | N>S | No |
ExAC gnomAD |
|
| rs1018357424 | 402 | H>P | No |
TOPMed gnomAD |
|
| rs1018357424 | 402 | H>R | No |
TOPMed gnomAD |
|
| rs1046334001 | 404 | S>C | No | TOPMed | |
| TCGA novel | 404 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs187305147 | 407 | L>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs757744938 | 411 | A>P | No |
ExAC gnomAD |
|
| rs2091285753 | 412 | K>T | No | TOPMed | |
| rs2125650951 | 415 | D>A | No | Ensembl | |
| rs2091285796 | 415 | D>N | No |
TOPMed gnomAD |
|
| rs2091285906 | 417 | P>S | No | gnomAD | |
| rs2091285988 | 422 | P>L | No |
TOPMed gnomAD |
|
| rs1172161441 | 423 | A>P | No | gnomAD | |
| rs1172161441 | 423 | A>T | No | gnomAD | |
|
rs1559472589 COSM1044844 COSM1044846 |
423 | A>V | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs539373016 | 424 | L>F | No |
1000Genomes ExAC gnomAD |
|
| rs1446572876 | 424 | L>H | No |
TOPMed gnomAD |
|
| rs1329757276 | 425 | S>C | No |
TOPMed gnomAD |
|
| rs1329757276 | 425 | S>Y | No |
TOPMed gnomAD |
|
| rs768354951 | 426 | T>A | No |
ExAC gnomAD |
|
| rs184123382 | 426 | T>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs184123382 | 426 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs768354951 | 426 | T>P | No |
ExAC gnomAD |
|
| rs769904264 | 427 | V>M | No |
ExAC gnomAD |
|
| rs759760181 | 428 | P>L | No |
ExAC gnomAD |
|
| rs773271563 | 428 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs2091286367 | 429 | P>S | No | Ensembl | |
| TCGA novel | 430 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374206608 | 430 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs2091286454 | 431 | S>G | No | Ensembl | |
| rs1575564299 | 433 | K>N | No | Ensembl | |
| rs1166983222 | 434 | P>A | No | TOPMed | |
| rs1255955990 | 435 | T>I | No |
TOPMed gnomAD |
|
| rs1575564308 | 435 | T>P | No | Ensembl | |
| rs989523499 | 436 | A>T | No |
TOPMed gnomAD |
|
| rs764400135 | 437 | S>N | No |
ExAC gnomAD |
|
| rs1475435045 | 437 | S>R | No | TOPMed | |
| rs1367633555 | 438 | G>R | No | Ensembl | |
| rs762069247 | 439 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs754276393 | 439 | R>W | No |
ExAC gnomAD |
|
| TCGA novel | 443 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757913103 | 443 | F>Y | No |
ExAC gnomAD |
|
| rs1408770998 | 444 | R>S | No |
TOPMed gnomAD |
|
| rs75950689 | 445 | V>G | No | Ensembl | |
| rs779486608 | 445 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs779486608 | 445 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 445 | V>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751198756 | 446 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs780688512 | 447 | E>D | No |
ExAC gnomAD |
|
| rs754696971 | 447 | E>K | No |
ExAC gnomAD |
|
| rs769711239 | 448 | D>E | No |
ExAC gnomAD |
|
| rs748036941 | 448 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs748036941 | 448 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs748036941 | 448 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1559472663 | 449 | E>K | No | Ensembl | |
| rs1276007232 | 455 | D>G | No | gnomAD | |
| rs750438882 | 456 | K>E | No | Ensembl | |
| rs772301139 | 456 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs575732643 | 456 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs575732643 | 456 | K>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs775600938 | 457 | K>N | No |
ExAC gnomAD |
|
|
COSM1044847 COSM1044849 |
457 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs760642858 | 458 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs760642858 | 458 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs2091287679 | 459 | M>I | No | gnomAD | |
| rs1477257719 | 459 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1263277808 | 459 | M>V | No |
TOPMed gnomAD |
|
|
COSM4117697 rs368288263 COSM4117696 |
460 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs762168922 | 461 | L>F | No |
ExAC gnomAD |
|
| rs2091287736 | 461 | L>R | No | TOPMed | |
| rs1241461580 | 462 | S>L | No |
TOPMed gnomAD |
|
| rs2091287777 | 463 | T>P | No | gnomAD | |
| rs765528463 | 465 | V>L | No |
ExAC gnomAD |
|
| rs765528463 | 465 | V>M | No |
ExAC gnomAD |
|
| rs2091287853 | 467 | L>V | No | 1000Genomes | |
| rs2125651199 | 467 | L>W | No | Ensembl | |
|
rs1400633567 COSM252842 COSM252843 |
468 | R>C | ovary breast [Cosmic] | No |
cosmic curated gnomAD |
|
COSM4728703 COSM4728702 rs371900831 |
469 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs763407333 | 469 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs765908830 | 476 | R>C | No | Ensembl | |
| rs375411982 | 476 | R>H | No |
ESP gnomAD |
|
| rs367685167 | 478 | T>P | No |
ESP ExAC |
|
| rs2091288163 | 479 | S>C | No | TOPMed | |
| rs1332576150 | 480 | R>G | No |
TOPMed gnomAD |
|
| rs1233638417 | 483 | A>V | No |
TOPMed gnomAD |
|
| rs777663687 | 485 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs2125651268 | 486 | L>P | No | Ensembl | |
| rs2091288366 | 487 | N>Y | No | gnomAD | |
| rs938530790 | 488 | Q>H | No | TOPMed | |
| TCGA novel | 494 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749269882 | 494 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs188648899 | 497 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs896927852 | 497 | D>G | No | TOPMed | |
| rs2091288629 | 502 | D>N | No | TOPMed | |
| rs2091288709 | 509 | L>F | No | TOPMed | |
| rs1471411247 | 511 | R>T | No | gnomAD | |
| TCGA novel | 513 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1423190 COSM1423191 |
515 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2091288843 | 515 | N>H | No | gnomAD | |
| rs1417985069 | 515 | N>S | No | gnomAD | |
| rs1288346098 | 516 | I>T | No | Ensembl | |
| rs2091288914 | 517 | A>P | No | Ensembl | |
|
COSM6164681 COSM6164680 |
519 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1459214141 | 519 | P>S | No | gnomAD | |
| rs77047396 | 520 | G>C | No | Ensembl | |
| rs77047396 | 520 | G>S | No | Ensembl | |
| rs1391671220 | 523 | H>P | No |
TOPMed gnomAD |
|
| rs1386427589 | 524 | K>R | No | gnomAD | |
| rs766970075 | 525 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs774717922 | 525 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs752425329 | 528 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs766880641 | 528 | R>W | No |
ExAC gnomAD |
|
| rs2091289292 | 531 | S>I | No | Ensembl | |
| rs865847130 | 534 | R>W | No |
TOPMed gnomAD |
|
| rs2091289427 | 535 | G>C | No | TOPMed | |
| COSM5101288 | 535 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2091289450 | 536 | S>F | No |
TOPMed gnomAD |
|
|
COSM4932495 COSM4932496 |
539 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM136340 rs1396607962 COSM136339 |
541 | S>L | skin [Cosmic] | No |
cosmic curated gnomAD |
| rs1409414729 | 547 | D>N | No | gnomAD | |
| rs192737915 | 551 | R>W | No |
1000Genomes ExAC gnomAD |
|
|
COSM1727858 COSM1727857 rs757110662 |
552 | R>Q | liver [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
rs2091289747 COSM3427606 COSM3427607 |
552 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs375426401 | 553 | R>Q | No |
ESP TOPMed gnomAD |
|
| rs778911222 | 553 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs745731005 | 554 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs745731005 | 554 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs2091289905 | 555 | D>N | No | gnomAD | |
| rs2091289944 | 556 | K>R | No | Ensembl | |
| rs201010640 | 557 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs770048995 | 558 | S>I | No |
ExAC gnomAD |
|
| rs749722160 | 559 | G>E | No |
ExAC gnomAD |
|
| rs1444483771 | 559 | G>R | No |
TOPMed gnomAD |
|
| rs2091290180 | 561 | T>I | No | gnomAD | |
| rs2091290153 | 561 | T>P | No | TOPMed | |
| rs2091290250 | 562 | Y>H | No | TOPMed | |
| rs1559472853 | 563 | S>C | No | gnomAD | |
| rs1263353928 | 564 | W>C | No | gnomAD | |
| rs2125651489 | 564 | W>R | No | Ensembl | |
| rs1559472863 | 566 | R>* | No | Ensembl | |
| TCGA novel | 566 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1423192 COSM1423193 |
567 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759911370 | 567 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs771134998 | 567 | R>W | No |
ExAC gnomAD |
|
| rs1245532158 | 569 | S>N | No | TOPMed | |
| rs2091290580 | 570 | S>N | No | TOPMed | |
| rs2125651505 | 570 | S>R | No | 1000Genomes | |
| rs775184271 | 571 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1350242857 | 576 | S>G | No | TOPMed | |
|
COSM3066625 rs753471554 COSM3066624 |
581 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
RCV001814769 rs2091290924 |
582 | G>D | No |
ClinVar Ensembl dbSNP |
|
| rs1171046157 | 582 | G>S | No | gnomAD | |
| rs2091290924 | 582 | G>V | No | Ensembl | |
|
TCGA novel rs1252161386 |
583 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
TOPMed gnomAD NCI-TCGA |
| rs2091290979 | 584 | S>G | No | Ensembl | |
| rs2091291004 | 584 | S>R | No | TOPMed | |
| rs2091291027 | 585 | K>R | No | Ensembl | |
| rs1463991486 | 587 | S>N | No | gnomAD | |
| rs1377521999 | 588 | N>D | No | gnomAD | |
| rs372506333 | 588 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1394996343 | 589 | A>G | No | gnomAD | |
| rs1309984689 | 590 | S>G | No |
TOPMed gnomAD |
|
| TCGA novel | 590 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779943111 | 591 | G>A | No |
ExAC gnomAD |
|
| rs2091291294 | 591 | G>R | No | TOPMed | |
| rs779943111 | 591 | G>V | No |
ExAC gnomAD |
|
| rs748283044 | 592 | G>W | No |
ExAC gnomAD |
|
| rs1575564796 | 593 | V>G | No | Ensembl | |
| rs777977613 | 595 | K>R | No |
ExAC gnomAD |
|
| rs2091291520 | 597 | S>G | No | TOPMed | |
| rs1483220264 | 601 | N>K | No |
TOPMed gnomAD |
|
| rs1257141619 | 601 | N>S | No | gnomAD | |
| rs779111154 | 602 | N>K | No |
ExAC gnomAD |
|
| rs771349870 | 602 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs927235415 | 605 | G>A | No |
TOPMed gnomAD |
|
| rs927235415 | 605 | G>E | No |
TOPMed gnomAD |
|
| rs746215927 | 606 | G>D | No |
ExAC gnomAD |
|
| rs1475992897 | 608 | P>A | No | gnomAD | |
| rs938478231 | 609 | S>P | No | Ensembl | |
| rs1193295272 | 610 | S>G | No |
TOPMed gnomAD |
|
| rs2091291916 | 610 | S>N | No | Ensembl | |
| rs1193295272 | 610 | S>R | No |
TOPMed gnomAD |
|
| rs2091292010 | 611 | G>D | No | Ensembl | |
|
rs368877591 VAR_041098 COSM20628 |
611 | G>S | ovary an ovarian mucinous carcinoma sample; somatic mutation [Cosmic, UniProt] | No |
cosmic curated UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
| rs776129453 | 612 | S>L | No |
ExAC gnomAD |
|
| rs375560589 | 613 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1575564931 | 615 | N>S | No | Ensembl | |
| rs1575564931 | 615 | N>T | No | Ensembl | |
| rs1048625849 | 616 | P>A | No |
TOPMed gnomAD |
|
| rs1048625849 | 616 | P>S | No |
TOPMed gnomAD |
|
| rs766369522 | 618 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs766369522 | 618 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs1039912962 | 620 | S>L | No | gnomAD | |
| rs777782793 | 621 | G>D | No |
ExAC gnomAD |
|
| rs2091292565 | 622 | T>A | No | TOPMed | |
| rs777728432 | 624 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs377641503 | 624 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs531886799 | 625 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs983920991 | 625 | R>H | No |
TOPMed gnomAD |
|
| rs772412582 | 626 | C>F | No |
ExAC gnomAD |
|
| rs1465684872 | 626 | C>W | No |
TOPMed gnomAD |
|
| rs772412582 | 626 | C>Y | No |
ExAC gnomAD |
|
| rs371111065 | 627 | A>T | No |
ESP TOPMed gnomAD |
|
| rs780716662 | 627 | A>V | No |
ExAC gnomAD |
|
| rs892633766 | 628 | G>S | No |
TOPMed gnomAD |
|
| rs1418974259 | 628 | G>V | No |
TOPMed gnomAD |
|
| rs768308943 | 630 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1267081451 | 631 | N>S | No | gnomAD | |
| rs2125651777 | 632 | S>F | No | Ensembl | |
| rs1234814847 | 633 | M>T | No | gnomAD | |
| rs200446000 | 633 | M>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1205389094 | 637 | S>A | No | TOPMed | |
| rs772911877 | 638 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs772911877 | 638 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM1642262 COSM1642263 rs762661633 |
638 | R>H | stomach [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs772911877 | 638 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs766105676 | 639 | S>G | No |
ExAC gnomAD |
|
| rs915281472 | 639 | S>R | No | TOPMed | |
| rs751520689 | 640 | A>T | No |
ExAC gnomAD |
|
| rs1323663298 | 641 | G>V | No | gnomAD | |
|
rs1488057856 COSM3767430 COSM3767429 |
644 | V>I | liver [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1177395325 | 645 | E>D | No |
TOPMed gnomAD |
|
| rs753918087 | 645 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1050536145 | 645 | E>K | No |
TOPMed gnomAD |
|
| rs757349076 | 646 | S>R | No |
ExAC gnomAD |
|
| rs2125651853 | 647 | L>V | No | Ensembl | |
| rs547452465 | 648 | K>E | No |
1000Genomes ExAC gnomAD |
|
| rs750599041 | 649 | L>R | No |
ExAC gnomAD |
|
| rs2091293776 | 649 | L>V | No | Ensembl | |
| rs202018563 | 650 | M>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs959916241 | 652 | L>F | No | Ensembl | |
| rs1460255892 | 653 | C>R | No |
TOPMed gnomAD |
|
| TCGA novel | 655 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1044850 COSM1044852 rs747341844 |
655 | G>S | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs755592027 | 656 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs781714956 | 656 | S>C | No |
ExAC gnomAD |
|
| rs755592027 | 656 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs918393086 | 657 | Q>H | No |
TOPMed gnomAD |
|
| rs1575565168 | 657 | Q>P | No | Ensembl | |
|
COSM1288114 COSM1288115 |
658 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1164602685 | 659 | H>P | No |
TOPMed gnomAD |
|
| rs1164602685 | 659 | H>R | No |
TOPMed gnomAD |
|
| rs747678664 | 661 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs769280193 | 662 | T>I | No |
ExAC gnomAD |
|
|
COSM1044853 COSM1044855 |
663 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2091294271 | 663 | K>R | No | Ensembl | |
| rs1439198099 | 665 | I>T | No | TOPMed | |
| rs539205148 | 666 | I>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs748924017 | 666 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs2091294502 | 669 | Q>* | No | Ensembl | |
| rs73831264 | 672 | L>F | No |
ExAC gnomAD |
|
| rs2091294614 | 672 | L>V | No | TOPMed | |
| rs761809581 | 673 | S>L | No |
ExAC gnomAD |
|
| rs766686409 | 675 | S>F | No |
ExAC gnomAD |
|
| rs755388340 | 676 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs755388340 | 676 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs748632098 | 676 | S>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 681 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777298284 | 682 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs909657099 | 682 | K>R | No |
TOPMed gnomAD |
|
| rs909657099 | 682 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs771136594 | 683 | S>C | No | TOPMed | |
| TCGA novel | 683 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs771136594 | 683 | S>Y | No | TOPMed | |
| rs868087272 | 684 | T>M | No |
TOPMed gnomAD |
|
| rs2091295366 | 685 | W>* | No | Ensembl | |
| rs1318419924 | 686 | K>T | No | gnomAD | |
| rs745732466 | 687 | M>I | No | ExAC | |
| rs2125652030 | 688 | C>G | No | Ensembl | |
| rs771836456 | 688 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs775309135 | 689 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1456749270 | 690 | S>C | No |
TOPMed gnomAD |
|
|
rs2091295549 COSM1044856 COSM1044858 |
691 | S>F | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs1236390386 | 694 | N>S | No | gnomAD | |
| rs1347280119 | 695 | A>V | No |
TOPMed gnomAD |
|
| rs763903055 | 696 | G>R | No |
ExAC gnomAD |
|
| rs559298185 | 697 | Q>K | No |
1000Genomes ExAC gnomAD |
|
| rs2125652075 | 698 | V>A | No | Ensembl | |
| rs1419268397 | 698 | V>F | No |
TOPMed gnomAD |
|
| rs1378552411 | 699 | P>A | No |
TOPMed gnomAD |
|
| TCGA novel | 699 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751763652 | 700 | A>E | No |
ExAC gnomAD |
|
|
COSM1485195 COSM1485196 |
700 | A>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1415547911 | 703 | G>D | No | gnomAD | |
| rs202212938 | 703 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1053127526 | 704 | I>M | No |
TOPMed gnomAD |
|
| rs2125652095 | 704 | I>V | No | Ensembl | |
| TCGA novel | 705 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 706 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1487398749 | 708 | S>C | No | TOPMed | |
| rs2091296175 | 709 | D>A | No | TOPMed | |
| rs756684829 | 709 | D>H | No |
ExAC gnomAD |
|
| rs1340876130 | 710 | H>N | No | gnomAD | |
| rs2125652117 | 711 | M>I | No | Ensembl | |
| rs373360457 | 711 | M>V | No |
ExAC TOPMed gnomAD |
|
|
COSM213509 rs753406949 |
713 | D>H | breast [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1189835869 | 714 | T>I | No | gnomAD | |
| rs1189835869 | 714 | T>N | No | gnomAD | |
| rs1189835869 | 714 | T>S | No | gnomAD | |
| rs756760223 | 715 | T>I | No |
ExAC gnomAD |
|
|
COSM111473 COSM111472 |
715 | T>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756760223 | 715 | T>S | No |
ExAC gnomAD |
|
|
COSM1423194 COSM1423195 rs200514916 |
720 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs201545457 COSM5085525 |
720 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1345166460 | 721 | I>L | No | gnomAD | |
| rs771792782 | 721 | I>M | No |
ExAC gnomAD |
|
| rs1345166460 | 721 | I>V | No | gnomAD | |
| TCGA novel | 722 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1263389138 | 723 | S>N | No | gnomAD | |
| rs1235925342 | 724 | K>R | No |
TOPMed gnomAD |
|
| rs1191400089 | 725 | N>K | No | gnomAD | |
| rs2091296757 | 725 | N>S | No | Ensembl | |
| rs746754643 | 726 | L>M | No |
ExAC gnomAD |
|
| rs1172572917 | 728 | N>S | No |
TOPMed gnomAD |
|
| TCGA novel | 729 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs572465919 COSM1423197 COSM1423196 |
730 | V>M | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
|
COSM1044861 COSM1044859 |
731 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1360088099 | 732 | Q>P | No |
TOPMed gnomAD |
|
| rs2091296990 | 733 | L>V | No |
TOPMed gnomAD |
|
| rs1332673516 | 735 | L>V | No | gnomAD | |
| rs568652187 | 737 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1286114978 | 740 | I>T | No | gnomAD | |
| rs1241628706 | 740 | I>V | No |
TOPMed gnomAD |
|
| rs1349434015 | 742 | V>L | No | gnomAD | |
| rs536095148 | 743 | N>D | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 744 | I>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369209633 | 746 | R>Q | No |
ESP ExAC gnomAD |
|
| rs753156157 | 746 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1198750333 | 748 | P>A | No |
TOPMed gnomAD |
|
|
VAR_041099 rs2125652224 COSM20626 |
748 | P>L | ovary an ovarian serous carcinoma sample; somatic mutation [Cosmic, UniProt] | No |
cosmic curated Ensembl UniProt |
| rs764733898 | 751 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA |
| rs756776738 | 753 | L>M | No |
ExAC TOPMed gnomAD |
|
|
COSM4117699 COSM4117698 |
753 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs373360520 | 753 | L>Q | No | ESP | |
| rs748346256 | 755 | A>T | No | TOPMed | |
| rs139836206 | 756 | S>P | No |
1000Genomes ExAC gnomAD |
|
| rs139836206 | 756 | S>T | No |
1000Genomes ExAC gnomAD |
|
| rs2091297736 | 757 | S>R | No |
TOPMed gnomAD |
|
| rs1194193824 | 757 | S>T | No |
TOPMed gnomAD |
|
| rs2091297764 | 758 | P>S | No | TOPMed | |
|
COSM4117701 COSM4117700 |
759 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1487829018 | 759 | A>V | No |
TOPMed gnomAD |
|
| rs758115013 | 761 | C>F | No |
ExAC gnomAD |
|
| rs779555770 | 763 | H>R | No |
ExAC gnomAD |
|
| rs1405686320 | 763 | H>Y | No | gnomAD | |
| rs2091297919 | 764 | V>I | No | TOPMed | |
| rs1448921885 | 765 | I>F | No |
TOPMed gnomAD |
|
|
COSM649 VAR_041100 |
765 | I>M | Variant assessed as Somatic; MODERATE impact. a breast pleomorphic lobular carcinoma sample; somatic mutation [NCI-TCGA, UniProt] | No |
NCI-TCGA Cosmic UniProt |
No associated diseases with Q9NRH2
4 regional properties for Q9NRH2
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| myeloid cell differentiation | The process in which a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte, megakaryocyte, thrombocyte, or erythrocyte lineages. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
56 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O14757 | CHEK1 | Serine/threonine-protein kinase Chk1 | Homo sapiens (Human) | EV |
| Q8IWQ3 | BRSK2 | Serine/threonine-protein kinase BRSK2 | Homo sapiens (Human) | PR |
| Q8TDC3 | BRSK1 | Serine/threonine-protein kinase BRSK1 | Homo sapiens (Human) | SS |
| Q14680 | MELK | Maternal embryonic leucine zipper kinase | Homo sapiens (Human) | EV |
| P57059 | SIK1 | Serine/threonine-protein kinase SIK1 | Homo sapiens (Human) | PR |
| Q9BXA7 | TSSK1B | Testis-specific serine/threonine-protein kinase 1 | Homo sapiens (Human) | PR |
| O60285 | NUAK1 | NUAK family SNF1-like kinase 1 | Homo sapiens (Human) | PR |
| Q13131 | PRKAA1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Homo sapiens (Human) | EV |
| P54646 | PRKAA2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Homo sapiens (Human) | EV |
| Q8LPZ7 | CPK3 | Calcium-dependent protein kinase 3 | Oryza sativa subsp. japonica (Rice) | SS |
| P53682 | CPK23 | Calcium-dependent protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| Q84SL0 | CPK20 | Calcium-dependent protein kinase 20 | Oryza sativa subsp. japonica (Rice) | SS |
| P53684 | CPK7 | Calcium-dependent protein kinase 7 | Oryza sativa subsp japonica (Rice) | PR |
| Q5VQQ5 | CPK2 | Calcium-dependent protein kinase 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q75GE8 | CPK8 | Calcium-dependent protein kinase 8 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6K968 | CPK6 | Calcium-dependent protein kinase 6 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6I5I8 | CPK16 | Calcium-dependent protein kinase 16 | Oryza sativa subsp. japonica (Rice) | SS |
| Q8LIG4 | CIPK3 | CBL-interacting protein kinase 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q852Q1 | OSK4 | Serine/threonine protein kinase OSK4 | Oryza sativa subsp. japonica (Rice) | SS |
| Q852Q2 | OSK1 | Serine/threonine protein kinase OSK1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6ZLP5 | CIPK23 | CBL-interacting protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAX3 | CIPK33 | CBL-interacting protein kinase 33 | Oryza sativa subsp japonica (Rice) | PR |
| Q2QY53 | CIPK32 | CBL-interacting protein kinase 32 | Oryza sativa subsp japonica (Rice) | PR |
| Q38872 | CPK6 | Calcium-dependent protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q06850 | CPK1 | Calcium-dependent protein kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38869 | CPK4 | Calcium-dependent protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q39016 | CPK11 | Calcium-dependent protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42479 | CPK3 | Calcium-dependent protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZV15 | CPK20 | Calcium-dependent protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M9V8 | CPK10 | Calcium-dependent protein kinase 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7XJR9 | CPK16 | Calcium-dependent protein kinase 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZSA4 | CPK27 | Calcium-dependent protein kinase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E9C0 | CPK34 | Calcium-dependent protein kinase 34 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SSF8 | CPK30 | Calcium-dependent protein kinase 30 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38870 | CPK2 | Calcium-dependent protein kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42396 | CPK12 | Calcium-dependent protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SJ61 | CPK25 | Calcium-dependent protein kinase 25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FMP5 | CPK17 | Calcium-dependent protein kinase 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8W4I7 | CPK13 | Calcium-dependent protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIQ7 | CPK24 | Calcium-dependent protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LDI3 | CIPK24 | CBL-interacting serine/threonine-protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q93VD3 | CIPK23 | CBL-interacting serine/threonine-protein kinase 23 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FJ54 | CIPK20 | CBL-interacting serine/threonine-protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O65554 | CIPK6 | CBL-interacting serine/threonine-protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22971 | CIPK13 | CBL-interacting serine/threonine-protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94CG0 | CIPK21 | CBL-interacting serine/threonine-protein kinase 21 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FLZ3 | KIN12 | SNF1-related protein kinase catalytic subunit alpha KIN12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1PE17 | CPK18 | Calcium-dependent protein kinase 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38873 | CPK7 | Calcium-dependent protein kinase 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93759 | GK-1 | Calcium-dependent protein kinase 14 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NLQ6 | CPK32 | Calcium-dependent protein kinase 32 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q2V452 | CIPK3 | CBL-interacting serine/threonine-protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FKW4 | CPK28 | Calcium-dependent protein kinase 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42438 | CPK8 | Calcium-dependent protein kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P92958 | KIN11 | SNF1-related protein kinase catalytic subunit alpha KIN11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAGFKRGYDG | KIAGLYDLDK | TLGRGHFAVV | KLARHVFTGE | KVAVKVIDKT | KLDTLATGHL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FQEVRCMKLV | QHPNIVRLYE | VIDTQTKLYL | ILELGDGGDM | FDYIMKHEEG | LNEDLAKKYF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AQIVHAISYC | HKLHVVHRDL | KPENVVFFEK | QGLVKLTDFG | FSNKFQPGKK | LTTSCGSLAY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SAPEILLGDE | YDAPAVDIWS | LGVILFMLVC | GQPPFQEAND | SETLTMIMDC | KYTVPSHVSK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ECKDLITRML | QRDPKRRASL | EEIENHPWLQ | GVDPSPATKY | NIPLVSYKNL | SEEEHNSIIQ |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RMVLGDIADR | DAIVEALETN | RYNHITATYF | LLAERILREK | QEKEIQTRSA | SPSNIKAQFR |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QSWPTKIDVP | QDLEDDLTAT | PLSHATVPQS | PARAADSVLN | GHRSKGLCDS | AKKDDLPELA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GPALSTVPPA | SLKPTASGRK | CLFRVEEDEE | EDEEDKKPMS | LSTQVVLRRK | PSVTNRLTSR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KSAPVLNQIF | EEGESDDEFD | MDENLPPKLS | RLKMNIASPG | TVHKRYHRRK | SQGRGSSCSS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SETSDDDSES | RRRLDKDSGF | TYSWHRRDSS | EGPPGSEGDG | GGQSKPSNAS | GGVDKASPSE |
| 610 | 620 | 630 | 640 | 650 | 660 |
| NNAGGGSPSS | GSGGNPTNTS | GTTRRCAGPS | NSMQLASRSA | GELVESLKLM | SLCLGSQLHG |
| 670 | 680 | 690 | 700 | 710 | 720 |
| STKYIIDPQN | GLSFSSVKVQ | EKSTWKMCIS | STGNAGQVPA | VGGIKFFSDH | MADTTTELER |
| 730 | 740 | 750 | 760 | ||
| IKSKNLKNNV | LQLPLCEKTI | SVNIQRNPKE | GLLCASSPAS | CCHVI |