Q9NRD5
Gene name |
PICK1 (PRKCABP) |
Protein name |
PRKCA-binding protein |
Names |
Protein interacting with C kinase 1, Protein kinase C-alpha-binding protein |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9463 |
EC number |
|
Protein Class |
ARFAPTIN-RELATED (PTHR12141) |
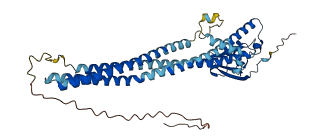
Descriptions
PRCKA-binding protein (PICK1) is a modular scaffold protein that regulates the activity and trafficking of several neural proteins. The membrane vesiculation capacity of the BAR domain is autoinhibited by PDZ and an acidic C-terminal tail (ACT). Upon deletion of ACT or PDZ domain, PICK1 clusters on vesicle-like structures that colocalize with the trans-Golgi network marker TGN38, driven by BAR domain. The ACT of one subunit of the PICK1 homodimer interacts with the PDZ and BAR domains of the other subunit. The autoinhibition is released by the binding of PDZ domain to receptor proteins or
Autoinhibitory domains (AIDs)
Target domain |
147-355 (BAR domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Structural analysis |
Target domain |
147-355 (BAR domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay |
Accessory elements
No accessory elements
References
- Karlsen ML et al. (2015) "Structure of Dimeric and Tetrameric Complexes of the BAR Domain Protein PICK1 Determined by Small-Angle X-Ray Scattering", Structure (London, England : 1993), 23, 1258-1270
- Madasu Y et al. (2015) "PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner", Molecular biology of the cell, 26, 1308-22
Autoinhibited structure

Activated structure

5 structures for Q9NRD5
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2GZV | X-ray | 112 A | A | 19-105 | PDB |
| 6AR4 | X-ray | 169 A | A/B | 1-105 | PDB |
| 6BJN | X-ray | 243 A | A/B | 1-105 | PDB |
| 6BJO | X-ray | 175 A | A/B | 1-105 | PDB |
| AF-Q9NRD5-F1 | Predicted | AlphaFoldDB |
311 variants for Q9NRD5
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1601933264 CA411507155 |
3 | A>V | No |
ClinGen Ensembl |
|
|
CA10228988 rs200693534 |
7 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 9 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1331717911 CA411507267 |
10 | E>K | No |
ClinGen gnomAD |
|
| TCGA novel | 11 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA324179862 rs757672073 |
12 | D>G | No |
ClinGen Ensembl |
|
|
rs758299374 CA10228990 |
13 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs777592756 CA10228991 |
14 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs746762898 CA10228992 |
14 | L>R | No |
ClinGen ExAC |
|
|
CA324180849 COSM3723048 rs889163499 |
15 | G>R | upper_aerodigestive_tract [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA411507817 rs1346856836 |
15 | G>V | No |
ClinGen TOPMed |
|
|
CA10229005 rs529594848 |
17 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA411507844 rs529594848 |
17 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1011662250 CA324180852 |
17 | P>S | No |
ClinGen Ensembl |
|
|
CA411507907 rs1343298477 |
22 | K>R | No |
ClinGen TOPMed |
|
|
rs1330468417 CA411507934 |
24 | T>S | No |
ClinGen TOPMed |
|
|
CA324180868 rs144230832 |
26 | Q>E | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA10229007 rs752748202 |
28 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA411507985 rs1267916199 |
28 | D>N | No |
ClinGen gnomAD |
|
|
CA10229008 rs758384023 |
29 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1376915390 CA411507996 |
29 | A>V | No |
ClinGen gnomAD |
|
|
rs5756896 CA10229009 |
30 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
| COSM3673279 | 31 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
CA10229011 rs375001291 |
33 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA411508020 rs1421037201 |
33 | I>N | No |
ClinGen gnomAD |
|
|
rs1375257084 CA411508026 |
34 | G>E | No |
ClinGen gnomAD |
|
|
rs1200286749 CA411508025 |
34 | G>R | No |
ClinGen TOPMed |
|
|
CA411508044 rs1393471162 |
36 | S>G | No |
ClinGen gnomAD |
|
|
rs914780868 CA324180888 |
37 | I>T | No |
ClinGen TOPMed |
|
|
CA411508081 rs1449682473 |
38 | G>E | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 42 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA411508162 rs1323301759 |
44 | C>R | No |
ClinGen gnomAD |
|
|
CA411508166 rs1327458589 |
44 | C>Y | No |
ClinGen gnomAD |
|
|
CA411508257 rs1276478116 |
50 | V>F | No |
ClinGen gnomAD |
|
|
rs1601944373 CA411509044 |
56 | T>P | No |
ClinGen Ensembl |
|
|
rs1277258113 CA411509048 |
56 | T>S | No |
ClinGen TOPMed |
|
|
CA324191387 rs373401475 |
59 | A>V | No |
ClinGen ESP TOPMed |
|
|
rs751472759 CA10229028 |
62 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs780886922 CA10229030 |
63 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs755889746 CA10229032 |
66 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs1360359047 CA411509164 |
68 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1450330922 CA411509181 |
69 | E>* | No |
ClinGen TOPMed |
|
| COSM3842672 | 69 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3554324 | 71 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
CA10229036 rs144498214 |
72 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs144498214 CA10229037 |
72 | G>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10229038 rs771440049 |
74 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1307843872 CA411509246 |
76 | R>T | No |
ClinGen gnomAD |
|
|
CA10229041 rs770073408 |
77 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA10229040 rs759886998 |
77 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs1306797006 CA411509256 |
78 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
COSM1416214 rs868326767 CA324191416 |
82 | T>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA Cosmic |
|
CA411509311 rs1601944546 |
84 | V>G | No |
ClinGen Ensembl |
|
|
CA411509305 rs763111317 |
84 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs763111317 CA10229043 |
84 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA324191438 rs908750329 |
86 | V>G | No |
ClinGen Ensembl |
|
|
CA10229044 rs764110971 |
87 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA324191490 rs767008199 |
89 | M>V | No |
ClinGen Ensembl |
|
|
CA324191497 rs1043621768 |
93 | V>A | No |
ClinGen Ensembl |
|
|
rs768613937 CA10229046 |
93 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1353828266 CA411509726 |
95 | G>A | No |
ClinGen gnomAD |
|
| COSM3554326 | 96 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs774355038 CA411509735 |
96 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1601948536 CA411509741 |
97 | V>G | No |
ClinGen Ensembl |
|
|
rs767430648 CA10229064 |
98 | T>P | No |
ClinGen ExAC gnomAD |
|
|
CA10229066 rs760644693 |
100 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA411509756 rs1278380187 |
100 | H>Y | No |
ClinGen gnomAD |
|
|
CA411509767 rs1256921063 |
101 | Y>* | No |
ClinGen gnomAD |
|
|
CA10229068 rs374422539 |
102 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs749368799 CA324193551 |
103 | K>E | No |
ClinGen Ensembl |
|
|
rs1224509124 CA411509790 |
105 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1361186118 CA411509795 |
105 | Q>H | No |
ClinGen TOPMed |
|
|
CA411509800 rs1245485885 |
106 | A>V | No |
ClinGen gnomAD |
|
|
CA10229069 rs754638692 |
107 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1476989308 CA411509810 |
108 | P>A | No |
ClinGen gnomAD |
|
|
rs200039533 CA324193578 |
112 | M>I | No |
ClinGen Ensembl |
|
|
rs1414699852 CA411509854 |
114 | L>P | No |
ClinGen gnomAD |
|
|
rs1197812788 CA411510213 |
118 | L>M | No |
ClinGen TOPMed |
|
| COSM6162288 | 123 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
CA324194730 rs1011013513 |
124 | R>Q | No |
ClinGen TOPMed |
|
|
rs1489069613 CA411510334 |
124 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA411510364 rs773477637 |
126 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA10229103 rs773477637 |
126 | V>G | No |
ClinGen ExAC gnomAD |
|
| COSM5749697 | 126 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs771064347 CA10229105 |
127 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs776641391 CA10229106 |
128 | N>D | No |
ClinGen ExAC |
|
|
rs769660774 CA10229108 |
129 | M>L | No |
ClinGen ExAC |
|
|
CA411510446 rs1172240986 |
130 | S>T | No |
ClinGen gnomAD |
|
|
CA411510477 rs1194766363 |
132 | G>R | No |
ClinGen gnomAD |
|
|
rs763721808 CA10229111 |
134 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs751138380 CA411510525 |
135 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10229112 rs751138380 |
135 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs766956739 CA10229114 |
136 | A>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 139 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs755404092 CA10229116 |
140 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs779380103 CA10229117 |
141 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs529753389 CA324194839 |
141 | R>W | No |
ClinGen 1000Genomes gnomAD |
|
|
CA10229118 rs752984261 |
143 | I>L | No |
ClinGen ExAC gnomAD |
|
|
CA411510683 rs1477970983 |
146 | N>S | No |
ClinGen gnomAD |
|
|
rs1415020764 CA411511525 |
151 | K>N | No |
ClinGen TOPMed |
|
| COSM6162287 | 152 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 154 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 157 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10229142 rs141358050 |
158 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs781381422 CA10229141 |
158 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1460371188 CA411511642 |
160 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA411511653 rs1183452291 |
161 | E>* | No |
ClinGen gnomAD |
|
|
rs974202812 CA324197375 |
165 | G>A | No |
ClinGen TOPMed |
|
|
CA411511845 rs1222342185 |
166 | M>L | No |
ClinGen gnomAD |
|
|
CA411511843 rs1222342185 |
166 | M>V | No |
ClinGen gnomAD |
|
|
CA10229173 rs766756193 |
167 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA411511884 rs1251254966 |
168 | E>D | No |
ClinGen gnomAD |
|
|
CA411511870 rs1202954419 |
168 | E>K | No |
ClinGen gnomAD |
|
|
rs1301431475 CA411511891 |
169 | H>Y | No |
ClinGen TOPMed |
|
|
CA411511925 rs1455635289 |
171 | K>N | No |
ClinGen TOPMed |
|
|
CA10229176 rs762087441 |
174 | L>R | No |
ClinGen ExAC gnomAD |
|
|
rs919508841 CA324197403 |
175 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA10229177 rs369159661 |
175 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA324197406 rs985113091 |
176 | A>V | No |
ClinGen TOPMed |
|
|
CA411511993 rs1408284648 |
179 | E>K | No |
ClinGen gnomAD |
|
|
CA411512011 rs1353885580 |
181 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs753896605 CA10229181 COSM3363639 |
182 | Q>L | kidney [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA411512024 rs1343231969 |
183 | T>I | No |
ClinGen gnomAD |
|
|
CA10229182 rs754970431 |
184 | H>D | No |
ClinGen ExAC gnomAD |
|
| COSM1130434 | 184 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs140773654 CA10229186 |
185 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM1416215 CA10229185 rs199868329 |
185 | R>W | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
CA10229208 rs745458781 |
189 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA10229210 rs370494530 |
190 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA324197843 rs950072445 |
191 | F>L | No |
ClinGen Ensembl |
|
|
rs201758961 CA411512092 |
193 | V>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA10229212 rs201758961 |
193 | V>M | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA411512102 rs761099651 |
194 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10229213 rs773510787 |
194 | I>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 196 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10229218 rs138517790 |
197 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs776855622 CA10229217 |
197 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA411512125 rs1447947465 |
198 | E>D | No |
ClinGen gnomAD |
|
|
CA10229220 rs373829961 |
200 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA411512135 rs1223501775 |
200 | Q>R | No |
ClinGen gnomAD |
|
|
CA411512143 rs1388700038 |
201 | P>R | No |
ClinGen TOPMed |
|
|
rs1279616473 CA411512156 |
203 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA10229222 rs141463477 |
204 | S>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA411512161 rs141463477 |
204 | S>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM580036 rs200906170 CA324197905 |
205 | E>K | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen NCI-TCGA Cosmic cosmic curated 1000Genomes NCI-TCGA TOPMed gnomAD |
|
rs1025987976 CA324197911 |
206 | A>T | No |
ClinGen Ensembl |
|
| COSM3554327 | 209 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs767370449 CA10229224 |
209 | K>E | No |
ClinGen ExAC gnomAD |
|
| COSM3554328 | 210 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs755797154 CA10229226 |
211 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs748819803 CA10229228 |
212 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10229229 rs758898232 |
215 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778351060 CA10229230 |
215 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA411512249 rs151027653 |
217 | I>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA10229232 rs149406206 COSM388727 |
218 | E>K | lung [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA10229233 rs149406206 |
218 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA411512272 rs373937870 |
220 | F>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA324197963 rs974961347 |
221 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA10229237 COSM1220527 rs763096477 |
223 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs368390981 CA10229236 |
223 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA411512318 rs761803705 |
228 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774366608 CA10229239 |
228 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs767460364 CA10229241 |
229 | K>E | No |
ClinGen ExAC |
|
|
CA10229242 rs750223551 |
229 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755889770 CA10229243 |
230 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755889770 CA411512328 |
230 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA411512327 rs1359113183 |
230 | P>S | No |
ClinGen TOPMed |
|
|
rs1361263875 CA411512360 |
233 | T>M | No |
ClinGen gnomAD |
|
|
rs878967259 CA324198269 |
233 | T>P | No |
ClinGen Ensembl |
|
|
CA10229269 rs752849719 |
234 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10229268 rs550929865 |
234 | D>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs756522592 CA10229270 |
237 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1356135658 CA411512403 |
240 | N>S | No |
ClinGen gnomAD |
|
|
CA10229271 rs780240972 |
243 | I>N | No |
ClinGen ExAC gnomAD |
|
|
CA10229272 rs749565487 |
244 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA10229275 rs748234997 |
247 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA10229276 rs376594572 |
247 | R>H | No |
ClinGen ESP ExAC gnomAD |
|
|
CA10229279 rs770795862 |
250 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs1301712577 CA411512472 |
251 | K>R | No |
ClinGen gnomAD |
|
|
rs1601958645 CA411512480 |
252 | K>R | No |
ClinGen Ensembl |
|
|
rs999854781 CA324198327 |
255 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs199605777 CA10229282 COSM4939840 |
256 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| TCGA novel | 258 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs752277834 CA10229283 |
258 | F>S | No |
ClinGen ExAC gnomAD |
|
|
CA411512585 rs1373714380 |
266 | K>Q | No |
ClinGen gnomAD |
|
|
rs1280502953 CA411512599 |
268 | K>E | No |
ClinGen gnomAD |
|
|
rs914333902 CA324198810 |
271 | D>N | No |
ClinGen Ensembl |
|
|
COSM4104043 CA324198831 rs945697657 |
273 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA Cosmic Ensembl NCI-TCGA |
| TCGA novel | 276 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs747734248 CA10229305 |
277 | C>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs376840369 CA10229306 |
278 | I>T | No |
ClinGen ESP ExAC gnomAD |
|
|
CA411512704 rs1436116403 |
281 | G>S | No |
ClinGen gnomAD |
|
|
rs1289821026 CA411512715 |
282 | E>D | No |
ClinGen gnomAD |
|
|
rs749077696 CA10229341 |
282 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs756192771 CA324199261 |
283 | P>H | No |
ClinGen Ensembl |
|
|
rs201690115 CA324199263 |
284 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA324199270 rs779309005 |
285 | Y>* | No |
ClinGen Ensembl |
|
|
CA411512728 rs1216575475 |
285 | Y>H | No |
ClinGen gnomAD |
|
|
CA411512738 rs1312964485 |
286 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
CA411512737 rs1312964485 |
286 | R>P | No |
ClinGen TOPMed gnomAD |
|
|
rs768374079 CA10229342 |
286 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1601961139 CA411512743 |
287 | V>G | No |
ClinGen Ensembl |
|
|
rs748514389 CA324199278 |
287 | V>L | No |
ClinGen Ensembl |
|
|
CA10229345 rs771516897 |
288 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA411512747 rs1247371829 |
288 | S>I | No |
ClinGen gnomAD |
|
|
CA324199291 rs771516897 |
288 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs1165286735 CA411512754 |
289 | T>I | No |
ClinGen gnomAD |
|
|
CA411512750 rs772751539 |
289 | T>P | No |
ClinGen ExAC gnomAD |
|
|
rs772751539 CA10229346 |
289 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA324199324 rs1004271976 |
290 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1404463657 CA411512760 |
291 | N>H | No |
ClinGen gnomAD |
|
|
CA411512766 rs1346394372 |
291 | N>K | No |
ClinGen gnomAD |
|
|
CA411512764 rs1159382697 |
291 | N>S | No |
ClinGen gnomAD |
|
|
CA411512772 rs1320421578 |
292 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
CA411512771 rs1320421578 |
292 | Y>F | No |
ClinGen TOPMed gnomAD |
|
|
rs770416253 CA10229348 |
292 | Y>H | No |
ClinGen ExAC gnomAD |
|
| COSM4104044 | 299 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM392957 | 301 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
CA10229351 rs111975858 |
303 | E>A | No |
ClinGen ExAC gnomAD |
|
|
rs111975858 CA324199346 |
303 | E>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 304 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 304 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 305 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 306 | A>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1220526 CA10229352 rs373780505 |
306 | A>T | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA411512864 rs1224480074 |
306 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1286627775 CA411512898 |
311 | M>R | No |
ClinGen gnomAD |
|
|
CA411512907 rs1386481151 |
312 | R>H | No |
ClinGen TOPMed |
|
|
CA10229353 rs762062613 |
313 | K>N | No |
ClinGen ExAC TOPMed |
|
|
CA411512929 rs1249313746 |
316 | L>M | No |
ClinGen TOPMed |
|
|
CA10229354 rs767542796 |
316 | L>R | No |
ClinGen ExAC gnomAD |
|
|
CA411512937 rs1215836539 |
317 | E>G | No |
ClinGen TOPMed |
|
|
CA411512953 rs1267397284 |
319 | M>T | No |
ClinGen gnomAD |
|
|
CA10229357 rs779971688 |
320 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs754791623 CA10229359 |
323 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs7511294 CA324199379 |
323 | D>V | No |
ClinGen Ensembl |
|
|
rs778731511 CA10229360 |
324 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs376883577 CA324199397 |
326 | H>R | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1215524681 CA411513023 |
328 | Q>E | No |
ClinGen TOPMed |
|
|
CA324199838 rs915606766 |
328 | Q>H | No |
ClinGen Ensembl |
|
|
rs771625974 CA411513041 |
330 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs745323732 CA10229386 |
330 | I>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 331 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10229388 rs774938827 |
332 | F>C | No |
ClinGen ExAC gnomAD |
|
|
rs746669587 CA10229389 COSM1034092 |
336 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1269183909 CA411513078 |
336 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| COSM6095376 | 338 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs895122027 CA411513097 |
339 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs895122027 CA324199861 |
339 | S>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs200682835 CA10229391 |
340 | T>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1476062469 CA411513109 |
341 | M>I | No |
ClinGen gnomAD |
|
|
rs952399927 CA324199864 |
345 | Y>C | No |
ClinGen TOPMed |
|
|
rs773151921 CA10229394 |
347 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765117173 CA10229393 |
347 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs766481418 CA10229396 |
350 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA411513172 rs766481418 |
350 | A>T | No |
ClinGen ExAC gnomAD |
|
| COSM4104045 | 351 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs767894272 CA411513189 |
353 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767894272 CA10229399 |
353 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1227984224 CA411513188 |
353 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA411513193 rs1339750116 |
354 | D>Y | No |
ClinGen gnomAD |
|
|
rs868820760 CA324199900 |
355 | A>V | No |
ClinGen Ensembl |
|
|
CA10229400 rs753073612 |
356 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs756571367 CA10229401 |
357 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| TCGA novel | 358 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 361 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1447089053 CA411513238 |
361 | E>K | No |
ClinGen gnomAD |
|
|
CA411513239 rs1447089053 |
361 | E>Q | No |
ClinGen gnomAD |
|
|
rs749818794 CA10229404 |
362 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749818794 CA411513246 |
362 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779602161 CA10229406 |
365 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1387296537 CA411513281 |
367 | T>I | No |
ClinGen gnomAD |
|
|
CA411513279 rs1387296537 |
367 | T>N | No |
ClinGen gnomAD |
|
|
CA411513288 rs371575593 |
369 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs776294355 CA10229409 |
371 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA411513314 rs1285300109 |
373 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1320163856 CA411513326 |
374 | N>K | No |
ClinGen gnomAD |
|
|
CA411513329 rs1569205925 |
375 | Q>E | No |
ClinGen Ensembl |
|
|
CA411513331 rs769587321 |
375 | Q>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769587321 CA10229412 |
375 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA324199981 rs1005944442 |
376 | E>K | No |
ClinGen TOPMed |
|
|
CA411513368 rs1434064454 |
380 | D>G | No |
ClinGen gnomAD |
|
|
CA411513364 rs1321355978 |
380 | D>N | No |
ClinGen gnomAD |
|
|
rs1220485612 CA411513374 |
381 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA411513383 rs762939296 |
382 | E>D | No |
ClinGen ExAC TOPMed |
|
|
CA10229416 rs773063627 |
382 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA411513396 rs1242082785 |
384 | E>A | No |
ClinGen gnomAD |
|
|
CA10229419 rs774462263 |
386 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs923555440 CA324200028 |
386 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA411513419 rs1206416140 |
387 | E>G | No |
ClinGen gnomAD |
|
|
CA10229420 rs547766954 |
389 | D>H | No |
ClinGen 1000Genomes ExAC |
|
|
rs767801528 CA10229421 |
390 | T>K | No |
ClinGen ExAC gnomAD |
|
|
rs767801528 CA10229422 |
390 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs767801528 CA411513441 |
390 | T>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 391 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA411513448 rs1422449148 |
392 | A>T | No |
ClinGen gnomAD |
|
|
CA10229423 rs187753076 |
393 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA10229425 rs754213542 |
395 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA411513475 rs1386348153 |
396 | S>F | No |
ClinGen gnomAD |
|
|
rs1456707221 CA411513487 |
398 | D>H | No |
ClinGen gnomAD |
|
|
rs1337321149 CA411513498 |
399 | T>I | No |
ClinGen gnomAD |
|
|
CA324200079 rs766115790 |
400 | R>* | No |
ClinGen gnomAD |
|
|
CA411513501 rs1394100412 |
400 | R>P | No |
ClinGen gnomAD |
|
|
rs1049850068 CA324200081 |
401 | G>E | No |
ClinGen TOPMed |
|
|
CA411513503 rs1311527496 |
401 | G>R | No |
ClinGen gnomAD |
|
| COSM3554330 | 404 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1327869119 CA411513545 |
407 | D>E | No |
ClinGen gnomAD |
|
|
CA411513561 rs1265567981 |
410 | G>R | No |
ClinGen gnomAD |
|
|
CA411513583 rs1482458962 |
412 | W>* | No |
ClinGen gnomAD |
|
|
CA411513578 rs1224548222 |
412 | W>* | No |
ClinGen TOPMed |
|
|
CA411513596 rs1200282713 |
414 | D>G | No |
ClinGen gnomAD |
No associated diseases with Q9NRD5
No regional properties for Q9NRD5
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q9NRD5 | |||
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR12141 | ARFAPTIN-RELATED |
| PANTHER Subfamily | PTHR12141:SF1 | PRKCA-BINDING PROTEIN |
| PANTHER Protein Class | vesicle coat protein | |
| PANTHER Pathway Category |
Ionotropic glutamate receptor pathway PICK |
|
14 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endocytic vesicle membrane | The lipid bilayer surrounding an endocytic vesicle. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| postsynaptic early endosome | An early endosome of the postsynapse. It acts as the major sorting station on the endocytic pathway, targeting neurotransmitter receptors for degregation or recycling. |
| presynaptic membrane | A specialized area of membrane of the axon terminal that faces the plasma membrane of the neuron or muscle fiber with which the axon terminal establishes a synaptic junction; many synaptic junctions exhibit structural presynaptic characteristics, such as conical, electron-dense internal protrusions, that distinguish it from the remainder of the axon plasma membrane. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
| trans-Golgi network membrane | The lipid bilayer surrounding any of the compartments that make up the trans-Golgi network. |
10 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| Arp2/3 complex binding | Binding to an Arp2/3 complex, a protein complex that contains two actin-related proteins, Arp2 and Arp3, and five novel proteins (ARPC1-5). |
| G protein-coupled receptor binding | Binding to a G protein-coupled receptor. |
| identical protein binding | Binding to an identical protein or proteins. |
| membrane curvature sensor activity | Preferential binding of proteins on curved membranes. The binding to curved membranes by insertion (aka wedging) to curved membranes is mediated by both the hydrophobic and hydrophilic faces of the helix of membrane curvature sensing (MCS) proteins. |
| metal ion binding | Binding to a metal ion. |
| phospholipid binding | Binding to a phospholipid, a class of lipids containing phosphoric acid as a mono- or diester. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase C binding | Binding to protein kinase C. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
19 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to decreased oxygen levels | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting a decline in the level of oxygen. |
| cellular response to glucose starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of glucose. |
| dendritic spine maintenance | The organization process that preserves a dendritic spine in a stable functional or structural state. A dendritic spine is a specialized protrusion from a neuronal dendrite and is involved in synaptic transmission. |
| dendritic spine organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a dendritic spine. A dendritic spine is a specialized protrusion from a neuronal dendrite and is involved in synaptic transmission. |
| epigenetic regulation of embryonic gene expression | The covalent transfer of a methyl group to C-5 of cytosine that contributes to the epigenetic regulation of embryonic gene expression. |
| establishment of genomic imprinting | The establishment of epigenetic modifications (imprints) during gametogenesis, leading to an asymmetry between the maternal and paternal alleles, leading to differential expression of the corresponding alleles. This can happen through heterochromatin formation or through differential chromatin loop formation. |
| glial cell development | The process aimed at the progression of a glial cell over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell. |
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| long-term synaptic depression | A process that modulates synaptic plasticity such that synapses are changed resulting in the decrease in the rate, or frequency of synaptic transmission at the synapse. |
| monoamine transport | The directed movement of monoamines, organic compounds that contain one amino group that is connected to an aromatic ring by an ethylene group (-CH2-CH2-), into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| negative regulation of Arp2/3 complex-mediated actin nucleation | Any process that stops, prevents, or reduces the frequency, rate or extent of actin nucleation mediated by the Arp2/3 complex and interacting proteins. |
| neuronal ion channel clustering | The process in which voltage-gated ion channels become localized to distinct subcellular domains in the neuron. Specific targeting, clustering, and maintenance of these channels in their respective domains are essential to achieve high conduction velocities of action potential propagation. |
| positive regulation of receptor internalization | Any process that activates or increases the frequency, rate or extent of receptor internalization. |
| protein kinase C-activating G protein-coupled receptor signaling pathway | The series of molecular signals generated as a consequence of a G protein-coupled receptor binding to its physiological ligand, where the pathway proceeds with activation of protein kinase C (PKC). PKC is activated by second messengers including diacylglycerol (DAG). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| receptor clustering | The receptor metabolic process that results in grouping of a set of receptors at a cellular location, often to amplify the sensitivity of a signaling response. |
| regulation of Arp2/3 complex-mediated actin nucleation | Any process that modulates the frequency, rate or extent of actin nucleation mediated by the Arp2/3 complex and interacting proteins. |
| regulation of insulin secretion | Any process that modulates the frequency, rate or extent of the regulated release of insulin. |
| retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | The directed movement of substances from the Golgi back to the endoplasmic reticulum, mediated by vesicles bearing specific protein coats such as COPI or COG. |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MFADLDYDIE | EDKLGIPTVP | GKVTLQKDAQ | NLIGISIGGG | AQYCPCLYIV | QVFDNTPAAL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DGTVAAGDEI | TGVNGRSIKG | KTKVEVAKMI | QEVKGEVTIH | YNKLQADPKQ | GMSLDIVLKK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VKHRLVENMS | SGTADALGLS | RAILCNDGLV | KRLEELERTA | ELYKGMTEHT | KNLLRAFYEL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SQTHRAFGDV | FSVIGVREPQ | PAASEAFVKF | ADAHRSIEKF | GIRLLKTIKP | MLTDLNTYLN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KAIPDTRLTI | KKYLDVKFEY | LSYCLKVKEM | DDEEYSCIAL | GEPLYRVSTG | NYEYRLILRC |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RQEARARFSQ | MRKDVLEKME | LLDQKHVQDI | VFQLQRLVST | MSKYYNDCYA | VLRDADVFPI |
| 370 | 380 | 390 | 400 | 410 | |
| EVDLAHTTLA | YGLNQEEFTD | GEEEEEEEDT | AAGEPSRDTR | GAAGPLDKGG | SWCDS |