Q9NQT8
Gene name |
KIF13B (GAKIN, KIAA0639) |
Protein name |
Kinesin-like protein KIF13B |
Names |
Kinesin-like protein GAKIN |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:23303 |
EC number |
|
Protein Class |
STAR-RELATED LIPID TRANSFER PROTEIN 9 (PTHR47117) |
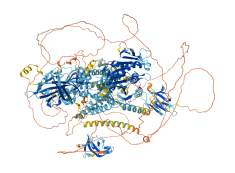
Descriptions
Kinesin-3 KIF13B autoinhibition involves the coiled-coil 1 (CC1) segment, which interacts with both the neck coil (NC) and the motor domain (MD). This interaction not only inhibits the NC-mediated dimerization, but also prevents the ADP release from the MD, effectively inhibiting the motor activity.
Autoinhibitory domains (AIDs)
Target domain |
1-389 (Motor domain and neck coil) |
Relief mechanism |
Partner binding |
Assay |
Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for Q9NQT8
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2COW | NMR | - | A | 1685-1771 | PDB |
| 3FM8 | X-ray | 230 A | A/B | 440-545 | PDB |
| 3GBJ | X-ray | 210 A | A/B/C | 4-352 | PDB |
| 3MDB | X-ray | 295 A | A/B | 440-545 | PDB |
| AF-Q9NQT8-F1 | Predicted | AlphaFoldDB |
1934 variants for Q9NQT8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1816746595 | 2 | G>E | No | TOPMed | |
| rs756089214 | 3 | D>A | No |
ExAC gnomAD |
|
| rs1816746185 | 3 | D>E | No | Ensembl | |
| rs756089214 | 3 | D>G | No |
ExAC gnomAD |
|
| rs763983325 | 3 | D>Y | No |
ExAC gnomAD |
|
| rs752467946 | 4 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs1180904272 | 4 | S>T | No | gnomAD | |
| rs752467946 | 4 | S>Y | No |
ExAC TOPMed gnomAD |
|
| rs767403615 | 5 | K>I | No |
ExAC gnomAD |
|
| rs767403615 | 5 | K>R | No |
ExAC gnomAD |
|
| rs1489156088 | 9 | A>V | No | gnomAD | |
| rs373241482 | 11 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1816745379 | 13 | R>G | No | Ensembl | |
| rs1816745285 | 13 | R>P | No | TOPMed | |
| rs765399690 | 16 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs765399690 | 16 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1816744835 | 17 | R>P | No | Ensembl | |
| rs1237673651 | 17 | R>W | No | gnomAD | |
| rs1030759553 | 18 | R>Q | No |
TOPMed gnomAD |
|
| rs1189249397 | 19 | E>D | No | gnomAD | |
| rs2130652102 | 20 | T>S | No | Ensembl | |
| TCGA novel | 21 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1018452157 | 22 | L>V | No |
TOPMed gnomAD |
|
| rs1210285664 | 23 | H>D | No |
TOPMed gnomAD |
|
| rs1199136655 | 23 | H>R | No |
TOPMed gnomAD |
|
| rs1210285664 | 23 | H>Y | No |
TOPMed gnomAD |
|
| rs755556447 | 25 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs747627167 | 26 | C>R | No |
ExAC gnomAD |
|
|
COSM1195198 COSM1195199 rs1212869754 |
26 | C>Y | lung [Cosmic] | No |
cosmic curated gnomAD |
| rs1338881074 | 27 | V>L | No | gnomAD | |
| rs573424392 | 28 | V>L | No | 1000Genomes | |
| rs781142652 | 29 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1563822917 | 30 | V>A | No | Ensembl | |
| rs1815978121 | 30 | V>L | No | TOPMed | |
| rs1380976838 | 31 | D>G | No | gnomAD | |
| rs1450506415 | 31 | D>H | No | gnomAD | |
| rs1336501112 | 32 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs754910588 | 33 | N>D | No |
ExAC gnomAD |
|
| rs1177291765 | 33 | N>K | No |
TOPMed gnomAD |
|
| rs1815976927 | 34 | K>R | No |
TOPMed gnomAD |
|
| rs1586980383 | 35 | V>G | No | Ensembl | |
| rs751345506 | 35 | V>I | No |
ExAC gnomAD |
|
| rs1413940192 | 37 | L>F | No | gnomAD | |
| rs952845164 | 39 | P>T | No | TOPMed | |
| rs373464930 | 42 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs373464930 | 42 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1400344030 | 43 | N>S | No | gnomAD | |
| rs1179025657 | 43 | N>Y | No |
TOPMed gnomAD |
|
| rs1458974369 | 44 | L>F | No | gnomAD | |
| rs1458974369 | 44 | L>I | No | gnomAD | |
| rs1815974542 | 44 | L>P | No | Ensembl | |
| rs1815974407 | 45 | S>T | No | Ensembl | |
| rs1311301305 | 47 | G>V | No | TOPMed | |
| rs1011102621 | 49 | A>T | No | gnomAD | |
| rs752753259 | 50 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs768039819 | 50 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs752753259 | 50 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1364457842 | 51 | G>D | No |
TOPMed gnomAD |
|
| rs2130380148 | 51 | G>S | No | Ensembl | |
| rs767555368 | 52 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1464629952 | 52 | Q>L | No |
TOPMed gnomAD |
|
|
COSM1648085 COSM750738 rs200573525 |
53 | P>L | lung [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs200573525 | 53 | P>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 55 | V>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs558541168 | 55 | V>M | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 56 | F>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 57 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs372924504 | 58 | Y>C | No |
ESP ExAC gnomAD |
|
| rs1384660537 | 59 | D>N | No | gnomAD | |
| rs1378202439 | 61 | C>F | No | gnomAD | |
| rs1334460577 | 64 | S>P | No | gnomAD | |
| rs538851654 | 65 | M>I | No |
1000Genomes ExAC gnomAD |
|
| rs762237959 | 65 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs1813157723 | 65 | M>T | No | Ensembl | |
| rs762237959 | 65 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1267078557 | 66 | D>G | No |
TOPMed gnomAD |
|
| rs1267078557 | 66 | D>V | No |
TOPMed gnomAD |
|
| rs199660413 | 67 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs965617359 | 68 | S>F | No | TOPMed | |
| rs376813155 | 68 | S>P | No |
ESP ExAC gnomAD |
|
| rs376813155 | 68 | S>T | No |
ESP ExAC gnomAD |
|
| rs965617359 | 68 | S>Y | No | TOPMed | |
| rs1371440953 | 69 | V>I | No | gnomAD | |
| rs772116769 | 70 | K>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 71 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1423800580 | 71 | E>G | No |
TOPMed gnomAD |
|
| rs1813156463 | 72 | K>E | No | gnomAD | |
| rs953039420 | 72 | K>R | No |
TOPMed gnomAD |
|
| rs866873741 | 74 | A>G | No | Ensembl | |
| rs866873741 | 74 | A>V | No | Ensembl | |
| rs755825639 | 75 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1813045195 | 77 | D>N | No |
TOPMed gnomAD |
|
| COSM282224 | 77 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767296475 | 78 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1404488870 | 79 | V>L | No |
TOPMed gnomAD |
|
| rs1813044782 | 80 | F>L | No | Ensembl | |
| rs759071302 | 81 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs751114531 | 81 | K>R | No |
ExAC gnomAD |
|
| rs751114531 | 81 | K>T | No |
ExAC gnomAD |
|
| rs1384749364 | 82 | C>R | No | gnomAD | |
| rs1350404173 | 84 | G>E | No | TOPMed | |
| rs1813043867 | 85 | E>K | No | gnomAD | |
| rs1203433257 | 86 | N>T | No |
TOPMed gnomAD |
|
| rs1426507325 | 88 | L>P | No | gnomAD | |
| rs772875337 | 91 | A>S | No |
ExAC gnomAD |
|
| rs772875337 | 91 | A>T | No |
ExAC gnomAD |
|
| rs1438867821 | 91 | A>V | No | Ensembl | |
| rs769508690 | 92 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs761606086 | 93 | D>E | No |
ExAC gnomAD |
|
| rs1243470898 | 93 | D>G | No |
TOPMed gnomAD |
|
| rs1243470898 | 93 | D>V | No |
TOPMed gnomAD |
|
| rs772311578 | 96 | N>D | No | ExAC | |
| rs745902872 | 97 | A>T | No |
ExAC gnomAD |
|
| rs1813041454 | 97 | A>V | No | Ensembl | |
| rs771331059 | 98 | C>G | No |
ExAC TOPMed gnomAD |
|
| rs1222963312 | 100 | F>I | No | gnomAD | |
| rs1190214308 | 101 | A>V | No | gnomAD | |
| rs201506710 | 102 | Y>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201506710 | 102 | Y>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs778148157 | 102 | Y>H | No |
ExAC gnomAD |
|
| rs1436255257 | 103 | G>* | No |
TOPMed gnomAD |
|
| rs371915765 | 103 | G>A | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 107 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1812933291 | 108 | G>* | No | Ensembl | |
| TCGA novel | 108 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748496343 | 112 | T>I | No |
ExAC gnomAD |
|
| rs781725176 | 113 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1442512766 | 114 | M>L | No |
TOPMed gnomAD |
|
|
COSM3834554 COSM3834555 |
114 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1243514772 | 115 | G>S | No | Ensembl | |
| rs1812932205 | 116 | T>P | No |
TOPMed gnomAD |
|
| rs1812931962 | 119 | Q>K | No | gnomAD | |
| rs1384412173 | 120 | P>T | No | gnomAD | |
| rs1812931695 | 121 | G>E | No | Ensembl | |
| rs1812931466 | 122 | L>I | No | TOPMed | |
| rs749154799 | 124 | P>R | No | TOPMed | |
| rs1471126219 | 128 | S>G | No | gnomAD | |
| rs746602364 | 128 | S>T | No |
ExAC gnomAD |
|
| rs143674632 | 131 | F>L | No |
1000Genomes ExAC gnomAD |
|
| rs1292738321 | 131 | F>L | No |
TOPMed gnomAD |
|
| rs749932898 | 132 | E>G | No |
ExAC gnomAD |
|
| rs1214134486 | 133 | R>* | No |
TOPMed gnomAD |
|
| rs757232762 | 133 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1586898251 | 134 | T>A | No | Ensembl | |
| rs1812928793 | 134 | T>I | No | Ensembl | |
| rs1218726419 | 135 | Q>E | No | TOPMed | |
| rs1812928419 | 135 | Q>H | No | Ensembl | |
| rs1812927978 | 137 | E>G | No | TOPMed | |
| rs540759947 | 137 | E>K | No | gnomAD | |
| rs763946380 | 138 | E>D | No |
ExAC gnomAD |
|
| rs2130315286 | 138 | E>K | No | Ensembl | |
| rs1586898181 | 142 | Q>K | No | Ensembl | |
| rs1812926967 | 146 | V>I | No | Ensembl | |
| rs36011290 | 147 | E>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1812926470 | 149 | S>A | No | TOPMed | |
| rs770405953 | 150 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs149530420 | 151 | M>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs149530420 | 151 | M>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs759625861 | 152 | E>K | No |
ExAC gnomAD |
|
| rs747353276 | 154 | Y>H | No |
ExAC gnomAD |
|
| rs1563769591 | 156 | E>D | No | Ensembl | |
|
COSM3648130 rs779713009 COSM3648131 |
159 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC TOPMed gnomAD |
| rs758026490 | 159 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs745358862 | 160 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs778450352 | 163 | D>N | No |
ExAC gnomAD |
|
| rs1024566514 | 166 | G>R | No | TOPMed | |
| rs759399782 | 167 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs377611918 | 167 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs759399782 | 167 | S>T | No |
ExAC TOPMed gnomAD |
|
|
rs1265643989 COSM3929534 COSM3929535 |
168 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs745585309 | 168 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1265643989 | 168 | R>S | No |
TOPMed gnomAD |
|
| rs1306106845 | 169 | Q>L | No |
TOPMed gnomAD |
|
| rs1306106845 | 169 | Q>R | No |
TOPMed gnomAD |
|
| rs778544096 | 170 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs748820334 | 171 | L>S | No |
ExAC gnomAD |
|
| rs777726670 | 173 | V>D | No |
ExAC gnomAD |
|
| rs1430534825 | 174 | R>T | No | gnomAD | |
| rs756050617 | 175 | E>D | No | ExAC | |
| rs374562840 | 176 | H>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs780911037 | 177 | S>G | No |
ExAC gnomAD |
|
| rs200153696 | 177 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs376293627 | 178 | V>M | No |
ESP TOPMed gnomAD |
|
| rs750613859 | 181 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs750613859 | 181 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs754007598 | 184 | D>A | No |
ExAC gnomAD |
|
| rs1206106036 | 184 | D>N | No |
TOPMed gnomAD |
|
| rs1210154058 | 185 | G>E | No |
TOPMed gnomAD |
|
| rs761142986 | 185 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs369434746 | 188 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1187814183 | 188 | K>I | No | TOPMed | |
|
COSM1331025 COSM1331024 |
189 | L>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1812729341 | 191 | V>I | No | TOPMed | |
| rs1430330848 | 192 | T>A | No |
TOPMed gnomAD |
|
| rs770602771 | 194 | Y>C | No |
ExAC gnomAD |
|
| rs1463651257 | 194 | Y>N | No | gnomAD | |
| rs1563765149 | 198 | E>G | No | Ensembl | |
| rs765997188 | 199 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1266413194 | 202 | S>F | No | gnomAD | |
| rs1300129201 | 205 | N>S | No |
TOPMed gnomAD |
|
| rs1209004480 | 207 | S>C | No | gnomAD | |
| rs769121640 | 208 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs747713433 | 208 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs747713433 | 208 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1812654517 | 214 | N>D | No | TOPMed | |
| rs1812654249 | 214 | N>K | No | Ensembl | |
| rs776390001 | 214 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1376470808 | 216 | N>K | No |
TOPMed gnomAD |
|
| rs116879347 | 217 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs746775301 | 219 | S>I | No |
ExAC gnomAD |
|
| rs780051811 | 220 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
|
rs757547963 COSM3996020 COSM3996019 |
221 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1237378889 | 221 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs749402531 | 224 | A>S | No |
ExAC gnomAD |
|
| rs1434174990 | 228 | I>V | No | gnomAD | |
| rs576265435 | 229 | T>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs756100272 | 229 | T>I | No |
ExAC gnomAD |
|
| rs767981563 | 230 | L>F | No |
ExAC gnomAD |
|
| rs767981563 | 230 | L>V | No |
ExAC gnomAD |
|
| rs1197762829 | 232 | H>R | No | gnomAD | |
| rs751911750 | 233 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs554021855 | 233 | T>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs554021855 | 233 | T>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751911750 | 233 | T>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 234 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1812650923 | 234 | L>V | No | Ensembl | |
| rs1203242454 | 235 | Y>C | No |
TOPMed gnomAD |
|
|
COSM245269 rs761192518 |
236 | D>N | prostate [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs761192518 | 236 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs913982292 | 237 | V>M | No |
TOPMed gnomAD |
|
| rs1812533598 | 242 | S>P | No | TOPMed | |
| rs865870469 | 243 | G>E | No | Ensembl | |
| COSM275730 | 244 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1812533212 | 245 | K>E | No | TOPMed | |
| rs764769996 | 247 | G>S | No | ExAC | |
| rs1812532937 | 248 | K>I | No | gnomAD | |
| rs1812532821 | 248 | K>N | No | gnomAD | |
| TCGA novel | 248 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761581932 | 251 | L>M | No |
ExAC gnomAD |
|
| rs1053435942 | 253 | D>A | No | Ensembl | |
| rs1276999381 | 255 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1319034727 | 257 | S>G | No |
TOPMed gnomAD |
|
| rs753393820 | 259 | R>* | No |
ExAC gnomAD |
|
| rs763630388 | 259 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs760274305 | 261 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs775520357 | 263 | T>I | No |
ExAC gnomAD |
|
| rs1812531068 | 264 | G>S | No | Ensembl | |
| rs933279686 | 265 | A>T | No |
TOPMed gnomAD |
|
| rs1586885606 | 266 | A>S | No | Ensembl | |
| rs759220461 | 268 | D>E | No |
ExAC gnomAD |
|
| rs1812530268 | 269 | R>K | No | Ensembl | |
| rs1337053152 | 271 | K>T | No | gnomAD | |
| rs1812529624 | 274 | S>R | No | Ensembl | |
| rs1812529497 | 275 | N>H | No | gnomAD | |
| rs774251931 | 275 | N>S | No |
ExAC gnomAD |
|
| rs1251545399 | 279 | S>F | No | TOPMed | |
| rs759577380 | 281 | T>A | No |
ExAC gnomAD |
|
| rs774015920 | 281 | T>I | No |
ExAC gnomAD |
|
| rs1472335140 | 282 | T>I | No | gnomAD | |
| rs770697789 | 282 | T>S | No |
ExAC gnomAD |
|
| rs1193179412 | 284 | G>S | No | gnomAD | |
| rs1487684645 | 285 | L>V | No |
TOPMed gnomAD |
|
| rs1044841485 | 287 | I>S | No | Ensembl | |
| rs1812468050 | 291 | A>S | No | TOPMed | |
| rs747064299 | 294 | S>N | No |
ExAC gnomAD |
|
| rs764782431 | 297 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs746318135 | 298 | N>K | No |
ExAC gnomAD |
|
|
COSM3432335 COSM261658 rs1302902650 |
299 | K>N | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated TOPMed gnomAD NCI-TCGA Cosmic NCI-TCGA |
| TCGA novel | 300 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1456539 COSM1456538 |
301 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779299462 | 303 | V>F | No |
ExAC gnomAD |
|
|
COSM3648124 rs1812466611 COSM3648125 |
304 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
|
COSM3925139 COSM3925138 |
306 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs375632161 | 306 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1812466352 | 308 | S>* | No | Ensembl | |
| rs1268938050 | 311 | T>S | No |
TOPMed gnomAD |
|
| rs1563762226 | 312 | W>* | No | Ensembl | |
| rs2130168577 | 316 | D>E | No | Ensembl | |
| rs376743977 | 316 | D>G | No | ESP | |
| rs1362407741 | 316 | D>N | No | TOPMed | |
| rs1206883853 | 318 | L>I | No | gnomAD | |
| rs747739598 | 318 | L>P | No |
ExAC gnomAD |
|
|
rs770147879 COSM3899496 COSM3899497 |
319 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed |
| rs770147879 | 319 | G>W | No |
ExAC TOPMed |
|
| rs1246522576 | 320 | G>D | No | gnomAD | |
| rs1246522576 | 320 | G>V | No | gnomAD | |
| TCGA novel | 320 | G>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766314108 | 321 | N>T | No |
ExAC gnomAD |
|
| rs1586871324 | 324 | T>P | No | Ensembl | |
| rs761608308 | 325 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs761608308 | 325 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs767663685 | 326 | M>I | No |
ExAC TOPMed gnomAD |
|
|
rs775729940 COSM4830292 COSM4830291 |
326 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC TOPMed gnomAD |
| rs775729940 | 326 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1455048993 | 328 | A>S | No | gnomAD | |
| rs1377863752 | 329 | T>A | No | TOPMed | |
| rs1411912384 | 330 | V>L | No |
TOPMed gnomAD |
|
| rs1411912384 | 330 | V>M | No |
TOPMed gnomAD |
|
| rs1028056429 | 332 | P>A | No | Ensembl | |
| rs1314506035 | 332 | P>L | No |
TOPMed gnomAD |
|
| rs142831178 | 334 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs1227564854 | 335 | D>G | No | TOPMed | |
| rs140358824 | 335 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs376444669 | 338 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs770228294 | 338 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs773863869 | 338 | D>H | No | ExAC | |
| rs1812059257 | 339 | E>G | No | Ensembl | |
| rs746510390 | 341 | L>V | No |
ExAC TOPMed gnomAD |
|
|
COSM3834552 COSM3834553 |
342 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1183816207 | 342 | S>L | No | gnomAD | |
| rs1255481321 | 343 | T>A | No |
TOPMed gnomAD |
|
| rs1250683588 | 345 | R>Q | No |
TOPMed gnomAD |
|
| rs779494332 | 345 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs372174415 | 347 | A>G | No |
ESP TOPMed |
|
| rs372174415 | 347 | A>V | No |
ESP TOPMed |
|
|
COSM3648122 COSM3648123 |
349 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1249894225 COSM2789027 COSM2789026 |
349 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs757905032 | 350 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs757905032 | 350 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1183383553 | 351 | K>N | No | TOPMed | |
| rs1038006307 | 352 | H>R | No | Ensembl | |
| rs750300915 | 352 | H>Y | No |
ExAC gnomAD |
|
| rs778842588 | 353 | I>V | No |
ExAC gnomAD |
|
| rs753614333 | 356 | H>R | No |
ExAC gnomAD |
|
| rs1586871061 | 359 | V>M | No | Ensembl | |
| rs1003953580 | 360 | N>S | No |
TOPMed gnomAD |
|
| rs886909920 | 362 | D>V | No | Ensembl | |
| rs751656609 | 363 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1812055382 | 364 | N>D | No | Ensembl | |
| rs1812055251 | 364 | N>K | No | gnomAD | |
| rs375870077 | 365 | A>P | No | ESP | |
| rs773808991 | 366 | R>* | No |
ExAC gnomAD |
|
| rs773808991 | 366 | R>G | No |
ExAC gnomAD |
|
| rs371165328 | 366 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs371165328 | 366 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs371165328 | 366 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs761773190 | 369 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs117139027 | 369 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs768346320 | 370 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1812053704 | 371 | L>F | No | TOPMed | |
| rs375537864 | 372 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1265020735 | 372 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1563755626 | 376 | E>G | No | Ensembl | |
|
TCGA novel rs1812052781 |
379 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs777021212 | 379 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1272692789 | 380 | E>A | No |
TOPMed gnomAD |
|
| rs1212375165 | 380 | E>K | No | Ensembl | |
| rs771520619 | 381 | Q>E | No |
ExAC gnomAD |
|
| rs745412085 | 383 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1333119701 | 384 | K>I | No | gnomAD | |
| rs778791498 | 384 | K>Q | No |
ExAC gnomAD |
|
| rs2130166350 | 385 | A>G | No | Ensembl | |
|
rs771289632 COSM3648120 COSM3648121 |
387 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2130150960 | 388 | M>I | No | Ensembl | |
| rs1444242139 | 388 | M>V | No | gnomAD | |
| rs758747096 | 390 | S>P | No |
ExAC gnomAD |
|
| rs1811967931 | 390 | S>Y | No | Ensembl | |
| rs1811967793 | 391 | P>A | No | Ensembl | |
| rs750522143 | 391 | P>Q | No |
ExAC gnomAD |
|
| rs1419320462 | 393 | L>V | No | TOPMed | |
| rs145715439 | 394 | K>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145715439 | 394 | K>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs35542230 | 396 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs764691078 | 396 | R>W | No |
ExAC gnomAD |
|
| rs375939324 | 398 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs375939324 | 398 | E>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs773997871 | 399 | E>D | No |
ExAC gnomAD |
|
| TCGA novel | 401 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4835720 COSM4835719 |
401 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770290644 | 402 | K>R | No |
ExAC gnomAD |
|
| TCGA novel | 403 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1811962666 | 403 | L>V | No | Ensembl | |
| rs748854849 | 404 | I>M | No |
ExAC gnomAD |
|
| rs1811962390 | 404 | I>V | No | Ensembl | |
| rs1737639261 | 406 | E>Q | No | TOPMed | |
|
TCGA novel rs1811961893 |
407 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs772680018 | 407 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs369778342 | 408 | T>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs754821297 | 409 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs375476627 | 409 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1811960985 | 412 | E>K | No |
TOPMed gnomAD |
|
| rs1254318117 | 414 | K>R | No | gnomAD | |
| rs1811960759 | 415 | L>V | No | Ensembl | |
|
COSM605432 COSM605431 rs779216622 |
418 | T>M | lung Variant assessed as Somatic; MODERATE impact. endometrium [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2130149807 | 420 | E>K | No | Ensembl | |
| rs1321185670 | 424 | E>V | No | gnomAD | |
| rs765928491 | 425 | R>* | No |
ExAC gnomAD |
|
| rs1157129117 | 425 | R>Q | No | gnomAD | |
| rs1328983825 | 427 | K>* | No | gnomAD | |
| TCGA novel | 428 | Q>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs187145887 | 429 | L>V | No |
1000Genomes ExAC gnomAD |
|
| rs750045090 | 430 | E>* | No |
ExAC gnomAD |
|
| rs764779364 | 431 | S>N | No |
ExAC gnomAD |
|
| rs761426546 | 434 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1811745098 | 434 | I>V | No | TOPMed | |
| TCGA novel | 435 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1418551643 | 436 | L>F | No |
TOPMed gnomAD |
|
| rs1189805051 | 437 | Q>K | No | gnomAD | |
| rs1465816209 | 438 | S>A | No |
TOPMed gnomAD |
|
| rs776668264 | 439 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1312399225 | 443 | V>F | No |
TOPMed gnomAD |
|
| rs1312399225 | 443 | V>I | No |
TOPMed gnomAD |
|
| rs929676085 | 444 | G>E | No | Ensembl | |
| rs760522054 | 444 | G>R | No |
ExAC gnomAD |
|
| rs771053234 | 445 | D>E | No |
ExAC gnomAD |
|
| rs371585117 | 445 | D>H | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 446 | D>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1404402880 | 446 | D>G | No | TOPMed | |
| rs749461076 | 446 | D>N | No |
ExAC gnomAD |
|
| rs1318603914 | 447 | K>* | No |
TOPMed gnomAD |
|
| rs1318603914 | 447 | K>Q | No |
TOPMed gnomAD |
|
| rs368623779 | 448 | C>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1178539640 | 452 | N>T | No | gnomAD | |
| rs1413077789 | 453 | L>P | No | gnomAD | |
| TCGA novel | 457 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781643536 | 458 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs781643536 | 458 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs755531808 | 458 | A>V | No |
ExAC gnomAD |
|
| rs759191313 | 459 | L>N | No |
ExAC gnomAD |
|
| rs550665832 | 460 | N>S | No |
1000Genomes ExAC gnomAD |
|
|
COSM486374 COSM486373 |
461 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1346797390 | 464 | V>L | No | gnomAD | |
| TCGA novel | 466 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1290199822 | 466 | Y>S | No |
TOPMed gnomAD |
|
| rs962940284 | 469 | E>D | No | gnomAD | |
| rs1474616069 | 469 | E>G | No |
TOPMed gnomAD |
|
| rs776752918 | 470 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs983008837 | 472 | L>F | No | TOPMed | |
| rs1471452631 | 472 | L>W | No | gnomAD | |
| rs1249567604 | 473 | I>T | No | gnomAD | |
| rs866414739 | 476 | A>T | No | gnomAD | |
| rs1197090261 | 478 | S>C | No |
TOPMed gnomAD |
|
| rs1197090261 | 478 | S>F | No |
TOPMed gnomAD |
|
| rs951526869 | 480 | D>G | No | Ensembl | |
| rs1811496199 | 481 | I>T | No | Ensembl | |
| rs769151121 | 483 | L>P | No |
ExAC gnomAD |
|
| rs780443382 | 485 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1306717700 | 486 | M>I | No | gnomAD | |
| rs758745849 | 486 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs1334272215 | 486 | M>T | No | gnomAD | |
|
COSM1456536 COSM1456537 rs1811495025 |
487 | G>E | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs201186759 | 487 | G>R | No | Ensembl | |
| rs1811495025 | 487 | G>V | No | gnomAD | |
| rs746255816 | 488 | I>L | No |
ExAC gnomAD |
|
| rs1370317127 | 488 | I>T | No |
TOPMed gnomAD |
|
| rs746255816 | 488 | I>V | No |
ExAC gnomAD |
|
| TCGA novel | 489 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2130067277 | 489 | L>V | No | Ensembl | |
| rs554993674 | 490 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1373630003 | 491 | E>G | No |
TOPMed gnomAD |
|
| rs1811494260 | 491 | E>K | No | TOPMed | |
| rs370408716 | 493 | C>R | No | ESP | |
| rs958670025 | 495 | I>T | No |
TOPMed gnomAD |
|
| rs377395370 | 495 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs752417793 | 496 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs752417793 | 496 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs778864660 | 498 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs778864660 | 498 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs778864660 | 498 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs561198851 | 500 | E>K | No | gnomAD | |
| rs1265131840 | 501 | G>D | No | gnomAD | |
| rs1811492571 | 502 | Q>K | No | TOPMed | |
| rs373090410 | 502 | Q>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs373090410 | 502 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1811492091 | 503 | V>A | No |
TOPMed gnomAD |
|
| rs1468046176 | 503 | V>F | No | gnomAD | |
| rs1811491986 | 504 | M>T | No | TOPMed | |
| rs759346506 | 505 | L>M | No |
ExAC gnomAD |
|
| rs774081586 | 505 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs774081586 | 505 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1348781979 | 507 | P>T | No |
TOPMed gnomAD |
|
|
COSM1099014 COSM1099016 |
508 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1309829409 | 509 | K>R | No | TOPMed | |
| rs370108872 | 510 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs370108872 | 510 | N>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs377720146 | 512 | R>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs369227969 | 513 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1372580071 | 515 | V>I | No |
TOPMed gnomAD |
|
| rs1372580071 | 515 | V>L | No |
TOPMed gnomAD |
|
| rs1811243169 | 520 | V>A | No | gnomAD | |
| rs1811243308 | 520 | V>L | No | Ensembl | |
| rs1586846270 | 522 | S>G | No | Ensembl | |
| rs1255737364 | 523 | P>T | No | gnomAD | |
| rs754104012 | 524 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1811242630 | 525 | Q>E | No | Ensembl | |
| rs921440927 | 525 | Q>H | No |
TOPMed gnomAD |
|
| TCGA novel | 526 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764185795 | 527 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1811241581 | 528 | H>R | No | gnomAD | |
| rs1338102896 | 528 | H>Y | No | gnomAD | |
| rs772639218 | 530 | D>E | No |
ExAC gnomAD |
|
| rs775376576 | 530 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1353146685 | 530 | D>H | No |
TOPMed gnomAD |
|
|
COSM1099011 COSM1099013 |
530 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1353146685 | 530 | D>Y | No |
TOPMed gnomAD |
|
| rs760188606 | 532 | I>L | No |
ExAC gnomAD |
|
| rs1409293375 | 537 | N>S | No | gnomAD | |
| TCGA novel | 538 | H>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1174078500 | 543 | N>H | No | gnomAD | |
| rs1157157989 | 543 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1455719954 | 545 | P>L | No | gnomAD | |
| rs752960231 | 547 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs767552972 | 548 | K>N | No |
ExAC gnomAD |
|
| rs1247750926 | 548 | K>T | No | gnomAD | |
|
COSM1099008 COSM1099010 rs769614957 |
549 | K>N | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs960188435 | 549 | K>R | No |
TOPMed gnomAD |
|
| rs775037511 | 551 | A>E | No |
ExAC gnomAD |
|
|
rs771524087 COSM270767 |
553 | R>* | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
COSM750742 COSM750741 |
553 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763432345 | 553 | R>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 554 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1251615901 | 555 | D>G | No |
TOPMed gnomAD |
|
| rs1345486265 | 557 | D>E | No |
TOPMed gnomAD |
|
| rs1811172659 | 558 | Q>E | No | Ensembl | |
| rs1811172540 | 559 | D>Y | No |
TOPMed gnomAD |
|
| rs868554153 | 560 | P>H | No |
TOPMed gnomAD |
|
| rs868554153 | 560 | P>L | No |
TOPMed gnomAD |
|
| rs773388574 | 560 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1370242088 | 561 | S>C | No | gnomAD | |
| rs1586844103 | 562 | M>T | No | TOPMed | |
| rs1168037660 | 563 | K>M | No |
TOPMed gnomAD |
|
| rs1168037660 | 563 | K>R | No |
TOPMed gnomAD |
|
| rs747376298 | 564 | N>K | No |
ExAC TOPMed gnomAD |
|
|
COSM5005280 rs780881097 COSM5005279 |
565 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1465883481 | 566 | N>K | No | gnomAD | |
| rs200170374 | 567 | S>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200170374 | 567 | S>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs536296566 | 568 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs779992016 | 569 | E>K | No |
ExAC gnomAD |
|
| rs1426311880 | 572 | D>G | No | gnomAD | |
| rs1175988261 | 574 | D>N | No | gnomAD | |
| rs370864469 | 575 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1208589568 | 575 | G>V | No | gnomAD | |
| rs1486581387 | 576 | D>G | No | gnomAD | |
| rs891802246 | 577 | S>A | No | Ensembl | |
| rs1811169121 | 577 | S>F | No | Ensembl | |
| rs1052272064 | 578 | S>A | No | Ensembl | |
| rs1811168847 | 578 | S>F | No | TOPMed | |
| rs1811168734 | 579 | S>G | No | TOPMed | |
| rs1811168598 | 579 | S>I | No | TOPMed | |
| rs922422712 | 580 | E>G | No | TOPMed | |
| rs778641038 | 580 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1442719645 | 581 | V>L | No |
TOPMed gnomAD |
|
| rs1040905926 | 583 | S>G | No |
TOPMed gnomAD |
|
| rs757062041 | 584 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs74341557 | 585 | V>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2130001606 | 585 | V>I | No | Ensembl | |
| rs755138971 | 586 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1811166697 | 590 | E>K | No | TOPMed | |
| COSM294506 | 592 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1811166111 | 593 | Q>* | No | TOPMed | |
| rs748631046 | 593 | Q>R | No | Ensembl | |
| rs574974074 | 596 | V>G | No | 1000Genomes | |
| rs1280810583 | 597 | T>I | No |
TOPMed gnomAD |
|
| rs762211309 | 598 | M>T | No |
ExAC gnomAD |
|
| rs1429720609 | 600 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1024856511 | 600 | A>V | No | TOPMed | |
| rs1811164711 | 601 | L>V | No | Ensembl | |
| rs1354784855 | 602 | G>D | No | gnomAD | |
| rs1811164314 | 603 | S>N | No | TOPMed | |
| rs377611054 | 603 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs972978590 | 604 | N>S | No |
TOPMed gnomAD |
|
|
COSM1212359 COSM1212358 rs750470565 |
606 | P>L | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs758444220 | 606 | P>S | No |
ExAC gnomAD |
|
| rs1811114955 | 610 | I>M | No |
TOPMed gnomAD |
|
| rs1298125778 | 610 | I>V | No |
TOPMed gnomAD |
|
| rs6986672 | 614 | L>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1387021346 | 616 | Q>E | No |
TOPMed gnomAD |
|
| rs764501247 | 616 | Q>R | No |
ExAC gnomAD |
|
| rs761007310 | 617 | Q>K | No |
ExAC gnomAD |
|
| rs2129991609 | 618 | H>R | No | Ensembl | |
| rs1811113658 | 619 | E>G | No | TOPMed | |
| rs1412315553 | 623 | R>* | No | gnomAD | |
|
COSM4695903 rs1416549201 COSM4695902 |
623 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1811112968 | 626 | L>V | No | TOPMed | |
| rs1811112837 | 627 | E>D | No | TOPMed | |
|
rs769742692 COSM3899490 COSM3899489 |
628 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM1099004 rs752388020 COSM1099002 |
628 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs369593806 | 629 | Q>R | No |
ESP ExAC gnomAD |
|
| rs1334415136 | 632 | M>I | No |
TOPMed gnomAD |
|
| rs1211964013 | 632 | M>T | No | gnomAD | |
|
COSM1099001 COSM1098999 |
635 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs763012855 COSM1456534 COSM1456535 |
636 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 638 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1442279282 | 641 | R>Q | No |
TOPMed gnomAD |
|
|
COSM1331027 COSM1331026 rs1306830181 |
641 | R>W | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1373896845 | 642 | R>K | No | gnomAD | |
| TCGA novel | 643 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769757334 | 643 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1439571543 | 644 | L>V | No | gnomAD | |
| rs780450879 | 645 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs780450879 | 645 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1451286934 | 646 | P>S | No | gnomAD | |
| rs565462520 | 648 | K>R | No |
1000Genomes ExAC gnomAD |
|
| rs986235822 | 649 | Q>E | No | Ensembl | |
| rs779119570 | 649 | Q>H | No |
ExAC gnomAD |
|
| rs757272844 | 651 | C>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 652 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778223174 | 652 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs754394313 | 652 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1207121901 | 653 | S>I | No | gnomAD | |
| rs1207121901 | 653 | S>T | No | gnomAD | |
| rs756503292 | 654 | M>K | No |
ExAC TOPMed gnomAD |
|
| rs1485051821 | 654 | M>L | No | gnomAD | |
| rs1811108034 | 655 | D>Y | No |
TOPMed gnomAD |
|
| rs142971472 | 656 | R>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1460933106 | 658 | S>F | No | gnomAD | |
| rs767089325 | 658 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs1811107193 | 660 | H>R | No | gnomAD | |
| rs751166000 | 660 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs200956809 | 661 | S>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs200956809 | 661 | S>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs374533712 | 662 | P>L | No |
ESP TOPMed gnomAD |
|
| rs761718096 | 662 | P>S | No |
ExAC gnomAD |
|
| rs2129989972 | 663 | S>C | No | 1000Genomes | |
| rs768334452 | 664 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1373710154 | 665 | Q>E | No | gnomAD | |
| TCGA novel | 665 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1811105570 | 666 | Q>E | No |
TOPMed gnomAD |
|
| rs1811105444 | 666 | Q>H | No | Ensembl | |
| rs1195486165 | 667 | R>C | No | gnomAD | |
| rs1195486165 | 667 | R>G | No | gnomAD | |
| rs573311825 | 667 | R>H | No |
1000Genomes ExAC TOPMed |
|
| rs1426937926 | 668 | L>V | No | gnomAD | |
| rs1470472689 | 670 | Q>* | No | Ensembl | |
| rs1470472689 | 670 | Q>E | No | Ensembl | |
| rs1015411999 | 671 | W>* | No |
TOPMed gnomAD |
|
| rs2129989578 | 671 | W>R | No | 1000Genomes | |
| rs1261348356 | 672 | A>D | No |
TOPMed gnomAD |
|
| rs1261348356 | 672 | A>V | No |
TOPMed gnomAD |
|
| rs1811103792 | 673 | E>G | No | TOPMed | |
| rs1211991763 | 673 | E>K | No | gnomAD | |
| rs1008738684 | 676 | E>G | No |
TOPMed gnomAD |
|
| rs376936122 | 677 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs769858692 | 677 | A>V | No |
ExAC gnomAD |
|
| rs1470737351 | 678 | T>A | No |
TOPMed gnomAD |
|
| rs781525515 | 678 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs781525515 | 678 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1811066918 | 680 | N>D | No | gnomAD | |
| rs747292009 | 680 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1811066520 | 681 | N>D | No | gnomAD | |
| rs749799843 | 681 | N>I | No |
ExAC gnomAD |
|
| rs749799843 | 681 | N>T | No |
ExAC gnomAD |
|
| rs1811065984 | 682 | S>G | No | TOPMed | |
| rs1811065568 | 682 | S>N | No | Ensembl | |
| rs1811065442 | 683 | L>P | No | TOPMed | |
| rs750099215 | 684 | M>V | No |
ExAC gnomAD |
|
| rs2129982007 | 685 | R>K | No | Ensembl | |
| rs1224083379 | 688 | E>D | No |
TOPMed gnomAD |
|
| rs1291937669 | 690 | I>F | No |
TOPMed gnomAD |
|
| rs1291937669 | 690 | I>V | No |
TOPMed gnomAD |
|
| rs1352587020 | 693 | A>S | No |
TOPMed gnomAD |
|
| rs1352587020 | 693 | A>T | No |
TOPMed gnomAD |
|
| rs1811064256 | 693 | A>V | No | gnomAD | |
| rs1811064140 | 694 | N>S | No |
TOPMed gnomAD |
|
| rs1164361783 | 695 | L>Q | No | TOPMed | |
| rs1277630291 | 695 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs753856156 | 697 | V>A | No |
ExAC gnomAD |
|
| rs1326637296 | 697 | V>L | No |
TOPMed gnomAD |
|
| rs1811063114 | 698 | R>T | No | TOPMed | |
| rs1328066135 | 700 | A>S | No | gnomAD | |
| rs1327348639 | 703 | I>T | No |
TOPMed gnomAD |
|
| rs994825623 | 703 | I>V | No |
TOPMed gnomAD |
|
| rs760541731 | 704 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs760541731 | 704 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2129981270 | 706 | E>* | No | 1000Genomes | |
| rs373789822 | 708 | D>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs763228452 | 710 | R>I | No |
ExAC gnomAD |
|
| rs1396249124 | 712 | E>Q | No | gnomAD | |
| rs2129981146 | 712 | E>V | No | Ensembl | |
| rs1811061300 | 713 | Y>C | No | TOPMed | |
| rs1466767686 | 716 | T>I | No | gnomAD | |
| rs1586840335 | 716 | T>P | No | Ensembl | |
| TCGA novel | 717 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs188349891 | 719 | I>M | No |
1000Genomes TOPMed gnomAD |
|
| rs1357995758 | 720 | P>A | No | TOPMed | |
| rs1202956051 | 722 | S>F | No |
TOPMed gnomAD |
|
| rs1171545299 | 723 | S>C | No | gnomAD | |
| rs748192834 | 725 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1811059843 | 727 | N>S | No | TOPMed | |
| rs777224705 | 729 | K>* | No |
ExAC gnomAD |
|
| rs768990907 | 729 | K>R | No |
ExAC gnomAD |
|
| rs1352269446 | 730 | R>* | No | gnomAD | |
| rs765456431 | 730 | R>L | No |
ExAC gnomAD |
|
| rs765456431 | 730 | R>Q | No |
ExAC gnomAD |
|
| rs1419620661 | 731 | G>D | No | gnomAD | |
| rs370214797 | 732 | S>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs370214797 | 732 | S>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs1162476711 | 732 | S>P | No | gnomAD | |
| rs1810851081 | 734 | L>I | No | TOPMed | |
| rs1408546453 | 734 | L>P | No | gnomAD | |
| rs1810850665 | 737 | P>S | No | TOPMed | |
| rs764132350 | 739 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs760781867 | 741 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2129942732 | 744 | K>E | No | Ensembl | |
| rs1183560285 | 745 | G>R | No | TOPMed | |
| rs1810849415 | 746 | K>R | No | Ensembl | |
| TCGA novel | 747 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1444725991 | 749 | Q>R | No | gnomAD | |
| rs1248445948 | 750 | I>M | No | TOPMed | |
| rs772658959 | 752 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs375884523 | 752 | S>P | No |
ESP TOPMed gnomAD |
|
| rs867925689 | 753 | L>S | No | Ensembl | |
| rs774791887 | 755 | K>R | No |
ExAC gnomAD |
|
| rs372624834 | 759 | R>G | No |
ESP ExAC gnomAD |
|
| rs1382470110 | 759 | R>K | No | TOPMed | |
| rs772898747 | 759 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs79504420 | 762 | D>G | No | Ensembl | |
| rs1810846724 | 764 | R>G | No | TOPMed | |
|
COSM3432334 COSM3432333 |
765 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs769227570 | 766 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs769227570 | 766 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1457159509 | 768 | Q>H | No |
TOPMed gnomAD |
|
| rs544708178 | 768 | Q>R | No | 1000Genomes | |
| rs1364156216 | 769 | E>* | No | gnomAD | |
| rs1810845934 | 771 | K>E | No | TOPMed | |
| rs781126671 | 772 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs896960113 | 772 | E>V | No |
TOPMed gnomAD |
|
| rs1036829044 | 773 | C>F | No |
TOPMed gnomAD |
|
| rs1810845164 | 773 | C>W | No | Ensembl | |
|
COSM454417 COSM454416 |
774 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 775 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779885612 | 776 | D>G | No |
ExAC gnomAD |
|
| rs746809012 | 776 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1393656800 | 777 | N>D | No |
TOPMed gnomAD |
|
| rs758192831 | 777 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1810844281 | 778 | P>L | No | Ensembl | |
| rs1810844410 | 778 | P>S | No |
TOPMed gnomAD |
|
| rs1005077444 | 779 | V>A | No |
TOPMed gnomAD |
|
|
COSM750751 COSM750752 |
779 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 780 | I>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1326613326 | 781 | R>* | No |
TOPMed gnomAD |
|
| rs1326613326 | 781 | R>G | No |
TOPMed gnomAD |
|
| rs758163466 | 781 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs758163466 | 781 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1586832338 | 782 | S>P | No | TOPMed | |
| rs1339602148 | 783 | Y>C | No | gnomAD | |
| rs1810773988 | 784 | F>L | No | Ensembl | |
| rs1810774112 | 784 | F>V | No | TOPMed | |
| rs1810773861 | 785 | K>R | No | TOPMed | |
| rs756295667 | 786 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs367974470 | 786 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1810773386 | 787 | A>V | No | Ensembl | |
| rs1220345843 | 789 | P>L | No |
TOPMed gnomAD |
|
| rs868353564 | 789 | P>S | No | TOPMed | |
| rs868353564 | 789 | P>T | No | TOPMed | |
| rs1465623549 | 791 | Y>C | No |
TOPMed gnomAD |
|
| rs1465623549 | 791 | Y>S | No |
TOPMed gnomAD |
|
| rs752151726 | 795 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs766868111 | 796 | N>S | No |
ExAC gnomAD |
|
| rs763246326 | 797 | H>D | No |
ExAC TOPMed gnomAD |
|
| rs1249247433 | 797 | H>Q | No |
TOPMed gnomAD |
|
| rs763246326 | 797 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs926239473 | 799 | L>F | No |
TOPMed gnomAD |
|
| rs926239473 | 799 | L>V | No |
TOPMed gnomAD |
|
| rs764798594 | 800 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs566501978 | 800 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM3648115 COSM3648114 |
801 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 802 | V>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1254724509 | 802 | V>L | No | gnomAD | |
| rs200010537 | 804 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs760142586 | 807 | L>F | No |
ExAC gnomAD |
|
| rs771987927 | 808 | E>G | No |
ExAC gnomAD |
|
| rs1004803364 | 808 | E>K | No |
TOPMed gnomAD |
|
| rs778837454 | 810 | L>F | No |
ExAC gnomAD |
|
| rs371398948 | 810 | L>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs778837454 | 810 | L>V | No |
ExAC gnomAD |
|
| rs781532057 | 811 | F>C | No |
ExAC TOPMed gnomAD |
|
| rs748428969 | 811 | F>I | No |
ExAC gnomAD |
|
| rs781532057 | 811 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs751607607 | 812 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs755139179 | 812 | Y>H | No |
ExAC gnomAD |
|
| rs780435093 | 814 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1397404726 | 818 | Y>H | No |
TOPMed gnomAD |
|
| rs762137974 | 819 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs762137974 | 819 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1239457344 | 820 | V>I | No |
TOPMed gnomAD |
|
| rs987371367 | 821 | P>A | No | TOPMed | |
| rs760195704 | 822 | I>F | No |
ExAC gnomAD |
|
| rs760195704 | 822 | I>V | No |
ExAC gnomAD |
|
| rs1457019458 | 823 | I>V | No | gnomAD | |
| rs1810766903 | 824 | N>D | No | Ensembl | |
| rs1234840182 | 824 | N>S | No |
TOPMed gnomAD |
|
| TCGA novel | 826 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1563734402 | 826 | K>Q | No | Ensembl | |
| rs759577444 | 827 | G>E | No | ExAC | |
| rs1024169681 | 827 | G>R | No | gnomAD | |
| rs1810766063 | 828 | E>D | No |
TOPMed gnomAD |
|
|
COSM3648108 COSM3648109 |
832 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1810750265 | 833 | L>Q | No | Ensembl | |
| rs112481059 | 835 | V>L | No |
ESP TOPMed gnomAD |
|
| rs112481059 | 835 | V>M | No |
ESP TOPMed gnomAD |
|
| rs1586831592 | 837 | V>G | No | Ensembl | |
| rs1348577782 | 838 | M>I | No | gnomAD | |
| rs757698662 | 838 | M>L | No |
ExAC gnomAD |
|
| rs757698662 | 838 | M>V | No |
ExAC gnomAD |
|
|
COSM5482958 rs1279400074 COSM5482957 |
839 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1221058237 | 839 | R>L | No |
TOPMed gnomAD |
|
| rs1221058237 | 839 | R>Q | No |
TOPMed gnomAD |
|
|
COSM1098996 COSM1098998 |
840 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1810748710 | 840 | L>V | No | gnomAD | |
| rs1255385311 | 841 | S>R | No |
TOPMed gnomAD |
|
| rs749791838 | 841 | S>T | No |
ExAC gnomAD |
|
| rs778170049 | 842 | G>S | No | ExAC | |
| rs756413071 | 843 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1342764895 | 844 | V>A | No |
TOPMed gnomAD |
|
| rs1342764895 | 844 | V>D | No |
TOPMed gnomAD |
|
| rs752432739 | 844 | V>F | No |
ExAC gnomAD |
|
|
rs752432739 COSM321226 COSM321227 |
844 | V>I | lung [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1399905993 | 847 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1439544762 | 847 | R>S | No |
TOPMed gnomAD |
|
| TCGA novel | 848 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754537721 | 849 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1012214797 | 851 | G>D | No | gnomAD | |
| rs766419855 | 852 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1168141885 | 853 | E>D | No | gnomAD | |
| rs1810745683 | 853 | E>K | No | Ensembl | |
| rs1810745427 | 854 | V>A | No | gnomAD | |
|
rs1478637697 COSM1456532 COSM1456533 |
855 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA gnomAD |
| rs763048072 | 855 | A>V | No |
ExAC gnomAD |
|
| TCGA novel | 856 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs931892075 | 856 | E>G | No | TOPMed | |
| TCGA novel | 856 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs987749124 | 856 | E>Q | No | TOPMed | |
| rs1586831404 | 857 | V>G | No | Ensembl | |
| rs773265419 | 857 | V>I | No |
ExAC gnomAD |
|
| rs1239170118 | 861 | K>N | No |
TOPMed gnomAD |
|
|
TCGA novel rs1810743847 |
862 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs373472045 | 862 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1310535250 | 863 | T>I | No |
TOPMed gnomAD |
|
| rs775807277 | 864 | Q>R | No |
ExAC gnomAD |
|
| rs2129922084 | 865 | E>* | No | Ensembl | |
| rs745969894 | 866 | N>I | No |
ExAC gnomAD |
|
| rs1000402231 | 868 | L>P | No |
TOPMed gnomAD |
|
| rs770986907 | 871 | M>R | No |
ExAC gnomAD |
|
| rs770986907 | 871 | M>T | No |
ExAC gnomAD |
|
| rs1305124823 | 871 | M>V | No |
TOPMed gnomAD |
|
| COSM5071800 | 873 | K>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 875 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1369583776 | 875 | L>P | No |
TOPMed gnomAD |
|
| rs1810463818 | 877 | A>P | No |
TOPMed gnomAD |
|
| TCGA novel | 878 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1287397147 | 878 | T>S | No | gnomAD | |
| rs978594772 | 879 | G>A | No | Ensembl | |
| TCGA novel | 881 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3996017 rs1586823542 COSM3996018 |
882 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs1810463204 | 883 | H>D | No | Ensembl | |
| rs769047547 | 884 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs2129860800 | 887 | F>L | No | Ensembl | |
| rs1307965845 | 887 | F>V | No | gnomAD | |
| rs1386964469 | 889 | F>S | No | gnomAD | |
| rs2129860739 | 892 | Y>* | No | Ensembl | |
| rs779818888 | 893 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs758007504 | 893 | S>N | No |
ExAC gnomAD |
|
| rs749898567 | 894 | F>L | No |
ExAC gnomAD |
|
| rs749898567 | 894 | F>V | No |
ExAC gnomAD |
|
| rs778917526 | 898 | Q>R | No |
ExAC gnomAD |
|
| rs1160387859 | 900 | P>L | No |
TOPMed gnomAD |
|
| rs1810461484 | 901 | V>L | No | TOPMed | |
| rs1378537317 | 903 | V>A | No | gnomAD | |
| rs751813268 | 904 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 904 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766537790 | 908 | D>G | No |
ExAC gnomAD |
|
| rs1037741101 | 910 | S>F | No | TOPMed | |
| rs1810460281 | 911 | S>A | No | Ensembl | |
| rs1563729301 | 913 | S>* | No | Ensembl | |
| rs1810459797 | 913 | S>T | No | TOPMed | |
|
rs368397416 COSM1212368 COSM1212369 |
914 | V>I | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1810459163 | 916 | K>R | No | TOPMed | |
| rs1810458570 | 918 | P>L | No | TOPMed | |
| rs1810458726 | 918 | P>S | No | gnomAD | |
| rs747335455 | 919 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1418576149 | 920 | C>Y | No |
TOPMed gnomAD |
|
| rs1385957045 | 921 | M>I | No | gnomAD | |
| rs779868044 | 922 | V>I | No |
ExAC gnomAD |
|
|
COSM3834549 rs771787654 COSM3834548 |
926 | H>D | breast [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs771787654 | 926 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs745515201 | 927 | C>G | No |
ExAC TOPMed gnomAD |
|
| rs745515201 | 927 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs1810457317 | 928 | N>D | No | Ensembl | |
| rs1810457197 | 928 | N>S | No | TOPMed | |
| rs1372952252 | 929 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1426146299 | 930 | F>L | No | gnomAD | |
| rs774027573 | 930 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs774027573 | 930 | F>Y | No |
ExAC TOPMed gnomAD |
|
| rs1810388117 | 931 | S>F | No | gnomAD | |
| rs770382593 | 932 | V>F | No |
ExAC gnomAD |
|
| rs1810387596 | 933 | N>K | No | Ensembl | |
| rs1457894146 | 933 | N>T | No |
TOPMed gnomAD |
|
| rs777583082 | 934 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs755976005 | 934 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs777583082 | 934 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1810386787 | 936 | E>D | No | Ensembl | |
| rs781018631 | 936 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs781018631 | 936 | E>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM1098992 rs1310607769 COSM1098990 |
940 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1810386168 | 941 | H>Q | No | Ensembl | |
| rs765457282 | 941 | H>R | No |
ExAC gnomAD |
|
| rs750650455 | 941 | H>Y | No |
ExAC gnomAD |
|
| rs1333636217 | 942 | L>F | No | gnomAD | |
| rs757303778 | 943 | S>P | No |
ExAC gnomAD |
|
| rs1810385800 | 943 | S>Y | No | Ensembl | |
| rs764266402 | 944 | E>* | No |
ExAC TOPMed gnomAD |
|
|
COSM750756 rs764266402 COSM750755 |
944 | E>K | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1810385204 | 946 | A>P | No | Ensembl | |
| rs1474046979 | 946 | A>V | No |
TOPMed gnomAD |
|
| rs1810384547 | 948 | A>G | No | gnomAD | |
| rs145324154 | 948 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145324154 | 948 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1810384547 | 948 | A>V | No | gnomAD | |
| rs771358165 | 949 | I>V | No |
ExAC gnomAD |
|
| rs1436929424 | 951 | V>A | No | gnomAD | |
| rs1176377601 | 951 | V>I | No | gnomAD | |
| rs2129841951 | 954 | H>R | No | Ensembl | |
| rs772772694 | 954 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs769276006 | 956 | I>M | No |
ExAC gnomAD |
|
| rs1810383155 | 958 | D>E | No | gnomAD | |
| rs993755183 | 958 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs768469903 | 960 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs746767801 | 960 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs768469903 COSM1098987 COSM1098989 |
960 | R>W | endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1810382397 | 961 | K>E | No | Ensembl | |
| TCGA novel | 962 | N>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3899483 COSM3899484 |
962 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1810382008 | 963 | P>L | No | TOPMed | |
| rs757567413 | 963 | P>S | No |
ExAC gnomAD |
|
| rs12549991 | 964 | A>P | No |
ExAC TOPMed gnomAD |
|
|
COSM1098986 COSM184722 rs12549991 |
964 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1420894873 | 965 | L>V | No |
TOPMed gnomAD |
|
| rs1810381324 | 967 | D>G | No | TOPMed | |
| TCGA novel | 975 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756236026 | 976 | R>C | No |
ExAC TOPMed gnomAD |
|
|
COSM3899481 COSM3899482 rs887441990 |
976 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs768092145 | 979 | R>Q | No |
ExAC gnomAD |
|
|
COSM1489223 rs753292093 COSM1489222 |
979 | R>W | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs2129840889 | 981 | R>I | No | Ensembl | |
| rs1283011788 | 982 | W>* | No | gnomAD | |
| rs1563726520 | 982 | W>L | No | Ensembl | |
| rs577422742 | 983 | S>G | No |
1000Genomes ExAC gnomAD |
|
| rs752769205 | 983 | S>N | No |
ExAC gnomAD |
|
| rs2129822358 | 984 | E>K | No | Ensembl | |
| rs755573729 | 985 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs766812686 | 989 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs2129822081 | 991 | F>L | No | Ensembl | |
| rs1810302015 | 991 | F>S | No | gnomAD | |
| rs1213569642 | 992 | W>* | No | TOPMed | |
| rs1267528238 | 993 | V>A | No | TOPMed | |
| rs1404929614 | 993 | V>I | No | gnomAD | |
|
COSM4485858 COSM4485857 |
995 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1171074566 | 996 | L>F | No | gnomAD | |
| rs2129821899 | 997 | E>K | No | Ensembl | |
|
COSM750757 COSM750758 |
998 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1478682942 | 1000 | E>D | No | gnomAD | |
|
COSM1456528 COSM1456529 |
1001 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372420177 | 1001 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs764819784 | 1005 | C>* | No |
ExAC TOPMed gnomAD |
|
| rs1473122222 | 1005 | C>S | No | gnomAD | |
| rs776192212 | 1007 | V>I | No |
ExAC gnomAD |
|
| rs768040042 | 1008 | E>* | No |
ExAC gnomAD |
|
| rs1015645347 | 1008 | E>A | No |
TOPMed gnomAD |
|
| rs971474919 | 1010 | I>V | No | Ensembl | |
| rs368227895 | 1012 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1810299605 | 1013 | K>R | No | Ensembl | |
| TCGA novel | 1015 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1810299222 | 1016 | P>R | No |
TOPMed gnomAD |
|
| rs771936727 | 1016 | P>S | No |
ExAC gnomAD |
|
| rs1380592611 | 1017 | T>A | No |
TOPMed gnomAD |
|
| rs1285399633 | 1017 | T>I | No | gnomAD | |
| rs1380592611 | 1017 | T>P | No |
TOPMed gnomAD |
|
| rs1810298650 | 1018 | G>R | No | gnomAD | |
| rs770077106 | 1020 | I>V | No |
ExAC gnomAD |
|
|
COSM3648104 COSM3648105 |
1022 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754987193 | 1024 | R>Q | No |
ExAC gnomAD |
|
|
COSM1456526 rs781387599 COSM1456527 |
1024 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1025 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1810154625 | 1027 | Q>R | No | TOPMed | |
| rs1351491411 | 1028 | S>Y | No | gnomAD | |
| rs200834673 | 1029 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs186826032 | 1029 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs774358524 | 1031 | V>A | No |
ExAC gnomAD |
|
| rs1159382252 | 1032 | Q>K | No | gnomAD | |
| rs1586815058 | 1034 | E>A | No | Ensembl | |
|
COSM1098984 COSM1098982 |
1034 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs548563840 | 1034 | E>K | No |
TOPMed gnomAD |
|
| rs548563840 | 1034 | E>Q | No |
TOPMed gnomAD |
|
|
COSM4414562 COSM4414563 |
1038 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1810152634 | 1038 | V>M | No | Ensembl | |
| rs1810152513 | 1039 | Q>P | No |
TOPMed gnomAD |
|
| rs1489018645 | 1041 | S>T | No | gnomAD | |
| rs776681784 | 1044 | L>S | No |
ExAC gnomAD |
|
| rs1810151576 | 1048 | E>G | No | TOPMed | |
| rs1235714674 | 1049 | E>K | No | Ensembl | |
|
COSM4394040 COSM4394039 rs1241710070 |
1050 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs768808287 | 1052 | L>M | No |
ExAC gnomAD |
|
| rs1433090450 | 1054 | V>I | No | gnomAD | |
| rs1433090450 | 1054 | V>L | No | gnomAD | |
| rs1810150517 | 1056 | I>F | No | TOPMed | |
| rs780623353 | 1056 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs780623353 | 1056 | I>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1059 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758834666 | 1059 | V>I | No |
ExAC gnomAD |
|
| rs1810149694 | 1060 | K>N | No | TOPMed | |
| rs1231884995 | 1062 | R>T | No |
TOPMed gnomAD |
|
| rs1301109730 | 1063 | P>L | No |
TOPMed gnomAD |
|
| rs1810148569 | 1066 | A>G | No | TOPMed | |
| rs1810148716 | 1066 | A>T | No | TOPMed | |
| rs1810148569 | 1066 | A>V | No | TOPMed | |
| rs1810148431 | 1067 | P>S | No | TOPMed | |
| rs374281215 | 1068 | R>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs763608396 | 1070 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs905792000 | 1070 | H>Y | No | Ensembl | |
| TCGA novel | 1070 | H>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1021427927 | 1071 | E>A | No | gnomAD | |
| rs755667406 | 1071 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs2129787243 | 1072 | T>A | No | 1000Genomes | |
| rs1455588526 | 1074 | H>P | No |
TOPMed gnomAD |
|
| rs1810116158 | 1075 | E>K | No | Ensembl | |
| rs1810116042 | 1076 | E>K | No | Ensembl | |
| rs370981148 | 1080 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1810001483 | 1086 | R>* | No | Ensembl | |
| rs539457354 | 1086 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1423609197 | 1088 | L>* | No |
TOPMed gnomAD |
|
| rs1810000946 | 1089 | E>G | No | TOPMed | |
| rs1303443570 | 1090 | R>T | No | TOPMed | |
| rs771282128 | 1092 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs759564273 COSM1489220 COSM1489221 |
1092 | R>H | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1390034917 | 1094 | K>E | No | gnomAD | |
| rs2129754404 | 1094 | K>N | No | Ensembl | |
| rs1269338608 | 1097 | N>D | No | TOPMed | |
| rs1412642018 | 1100 | T>A | No |
TOPMed gnomAD |
|
| rs749799694 | 1100 | T>I | No |
ExAC gnomAD |
|
| rs749799694 | 1100 | T>K | No |
ExAC gnomAD |
|
| rs1809999076 | 1101 | K>E | No | TOPMed | |
| rs1563721309 | 1102 | R>C | No | Ensembl | |
|
rs778060352 COSM3929533 COSM3929532 |
1102 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs770170380 | 1103 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs747792577 | 1104 | E>A | No |
ExAC gnomAD |
|
| rs754553518 | 1109 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1008332168 | 1110 | L>F | No | Ensembl | |
| rs1437028786 | 1111 | Q>E | No |
TOPMed gnomAD |
|
| rs1437028786 | 1111 | Q>K | No |
TOPMed gnomAD |
|
|
COSM1098979 COSM1098981 |
1112 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780144243 | 1114 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs780144243 | 1114 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1809996992 | 1115 | S>N | No | Ensembl | |
| rs747569217 | 1117 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs947207603 COSM1212362 COSM1212363 |
1117 | R>H | large_intestine [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1203582120 | 1118 | D>G | No |
TOPMed gnomAD |
|
| TCGA novel | 1118 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs371574802 | 1119 | K>I | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4925717 COSM4925718 |
1122 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1243190732 | 1123 | D>A | No | TOPMed | |
| rs1344099166 | 1124 | A>G | No |
TOPMed gnomAD |
|
| rs1401204056 | 1125 | D>E | No | gnomAD | |
| rs745825145 | 1126 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs745825145 | 1126 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM3648103 rs778861935 COSM3648102 |
1126 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs778861935 | 1126 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs745825145 | 1126 | R>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1128 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369600599 | 1128 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs372335072 | 1129 | Q>H | No |
ESP TOPMed gnomAD |
|
| rs1354529304 | 1132 | E>D | No | gnomAD | |
| TCGA novel | 1132 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs979231279 | 1133 | M>T | No |
TOPMed gnomAD |
|
| rs755254258 | 1134 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM245270 rs186117987 |
1134 | R>W | prostate [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1020789846 | 1137 | L>I | No | gnomAD | |
| rs1020789846 | 1137 | L>V | No | gnomAD | |
| rs1191142704 | 1138 | T>A | No | gnomAD | |
|
rs766406923 COSM271341 COSM3698714 |
1143 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM3899479 rs763045827 COSM3899480 |
1143 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| TCGA novel | 1144 | V>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1563720626 | 1144 | V>M | No | Ensembl | |
| rs1809964509 | 1145 | M>V | No | gnomAD | |
| rs1215634040 | 1146 | V>F | No | gnomAD | |
| rs1364729620 | 1147 | P>S | No |
TOPMed gnomAD |
|
| rs1364729620 | 1147 | P>T | No |
TOPMed gnomAD |
|
| rs369903381 | 1149 | A>T | No | Ensembl | |
| rs1429423049 | 1151 | S>T | No | gnomAD | |
| rs1809963609 | 1152 | G>R | No | Ensembl | |
| rs1340647020 | 1153 | I>T | No | gnomAD | |
| rs1338277873 | 1154 | P>A | No |
TOPMed gnomAD |
|
| rs1467969961 | 1156 | A>S | No | gnomAD | |
| rs1405046468 | 1157 | P>A | No | gnomAD | |
|
TCGA novel rs1809962725 |
1158 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1452476271 | 1159 | E>G | No |
TOPMed gnomAD |
|
| rs1370900636 | 1161 | T>I | No |
TOPMed gnomAD |
|
| rs1370900636 | 1161 | T>N | No |
TOPMed gnomAD |
|
| rs1225170775 | 1161 | T>S | No | gnomAD | |
| TCGA novel | 1162 | P>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs373318729 | 1162 | P>S | No |
ESP TOPMed gnomAD |
|
| rs1809924947 | 1164 | P>R | No | TOPMed | |
| rs775020582 | 1166 | M>V | No |
ExAC TOPMed gnomAD |
|
| COSM271936 | 1167 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1160099694 | 1167 | E>D | No | gnomAD | |
| rs370420261 | 1167 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1809924201 | 1168 | T>R | No | TOPMed | |
| rs759109548 | 1169 | H>D | No |
ExAC TOPMed gnomAD |
|
| rs1406213196 | 1169 | H>Q | No |
TOPMed gnomAD |
|
| rs759109548 | 1169 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1433707066 | 1170 | I>V | No |
TOPMed gnomAD |
|
| TCGA novel | 1172 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773971616 | 1172 | V>I | No |
ExAC gnomAD |
|
| rs1481026194 | 1173 | I>T | No | gnomAD | |
| rs2129738317 | 1176 | D>E | No | Ensembl | |
| rs770410591 | 1176 | D>N | No |
ExAC gnomAD |
|
| rs761127372 | 1179 | A>D | No |
ExAC gnomAD |
|
| rs975836913 | 1180 | D>G | No |
TOPMed gnomAD |
|
| rs376733489 | 1180 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs376733489 | 1180 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1809732123 | 1181 | D>E | No |
TOPMed gnomAD |
|
| rs2129697783 | 1181 | D>N | No | Ensembl | |
| rs557959639 | 1182 | F>S | No | 1000Genomes | |
| rs1284836248 | 1183 | S>T | No |
TOPMed gnomAD |
|
| rs1269754390 | 1184 | S>P | No | gnomAD | |
| TCGA novel | 1190 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1320123538 | 1190 | D>N | No | gnomAD | |
| rs1408719031 | 1191 | P>L | No | gnomAD | |
| TCGA novel | 1191 | P>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773848274 | 1191 | P>S | No |
ExAC gnomAD |
|
| rs773848274 | 1191 | P>T | No |
ExAC gnomAD |
|
| rs1809730849 | 1193 | A>G | No | TOPMed | |
| rs1174784751 | 1193 | A>P | No |
TOPMed gnomAD |
|
| rs770463620 | 1195 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1426208158 | 1197 | D>H | No |
TOPMed gnomAD |
|
| rs1426208158 | 1197 | D>N | No |
TOPMed gnomAD |
|
| rs1278099723 | 1197 | D>V | No |
TOPMed gnomAD |
|
|
COSM1331028 COSM1331029 rs965661459 |
1198 | A>E | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs965661459 | 1198 | A>V | No | gnomAD | |
| rs1563717010 | 1202 | G>E | No | Ensembl | |
| rs1210728101 | 1204 | E>Q | No | TOPMed | |
| rs1461376417 | 1205 | E>G | No | gnomAD | |
| rs1462418137 | 1205 | E>K | No | gnomAD | |
| rs1809728358 | 1206 | E>* | No | TOPMed | |
| TCGA novel | 1206 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1643779177 | 1206 | E>G | No |
TOPMed gnomAD |
|
| rs1643779177 | 1206 | E>V | No |
TOPMed gnomAD |
|
| rs1809727921 | 1207 | E>D | No | Ensembl | |
| rs1239330846 | 1208 | F>I | No | TOPMed | |
| rs1239330846 | 1208 | F>V | No | TOPMed | |
| rs372213370 | 1209 | F>S | No |
ESP TOPMed |
|
| rs1007784849 | 1210 | E>D | No | gnomAD | |
| TCGA novel | 1210 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1809727178 | 1212 | Q>* | No | TOPMed | |
| rs768312001 | 1212 | Q>H | No |
ExAC gnomAD |
|
| rs746142189 | 1213 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1809726726 | 1214 | V>L | No | TOPMed | |
| rs1381110404 | 1215 | K>N | No | TOPMed | |
| rs778961887 | 1217 | H>D | No | ExAC | |
| rs1809726281 | 1217 | H>P | No | gnomAD | |
| rs1809726281 | 1217 | H>R | No | gnomAD | |
| rs753904519 | 1218 | D>A | No |
ExAC gnomAD |
|
| rs753904519 | 1218 | D>G | No |
ExAC gnomAD |
|
| rs1297012294 | 1219 | G>A | No | gnomAD | |
| rs1441937748 | 1219 | G>R | No |
TOPMed gnomAD |
|
| rs1017827737 | 1220 | E>V | No | TOPMed | |
| rs907831530 | 1221 | V>A | No |
TOPMed gnomAD |
|
| rs907831530 | 1221 | V>G | No |
TOPMed gnomAD |
|
| rs974232785 | 1221 | V>L | No |
TOPMed gnomAD |
|
| rs758101383 | 1224 | E>* | No |
ExAC gnomAD |
|
| rs750054842 | 1225 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs750054842 | 1225 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs764108568 | 1226 | S>F | No |
TOPMed gnomAD |
|
| rs1211347611 | 1226 | S>P | No |
TOPMed gnomAD |
|
| rs1563715424 | 1227 | W>* | No | Ensembl | |
| rs1809633383 | 1228 | D>V | No | Ensembl | |
| rs776651773 | 1230 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs367974170 | 1230 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1273670588 | 1233 | G>S | No | gnomAD | |
| rs749564397 | 1236 | Q>* | No |
ExAC TOPMed gnomAD |
|
|
COSM3648100 COSM3648101 |
1236 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1809632056 | 1237 | L>R | No | TOPMed | |
| rs773532755 | 1239 | R>K | No |
ExAC gnomAD |
|
| rs769943486 | 1239 | R>S | No |
ExAC gnomAD |
|
| rs1223678476 | 1240 | G>V | No | gnomAD | |
| rs748115588 | 1241 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1809630818 | 1242 | P>H | No | Ensembl | |
| rs755501198 | 1242 | P>S | No |
ExAC gnomAD |
|
| rs2129674259 | 1243 | V>A | No | Ensembl | |
| rs780444269 | 1243 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1809630277 | 1244 | D>Y | No | gnomAD | |
| rs750106779 | 1245 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs1286631144 | 1245 | E>G | No | gnomAD | |
| rs750106779 | 1245 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs756797507 | 1246 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs192711591 | 1246 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1469015622 | 1247 | L>F | No | TOPMed | |
| rs1336922072 | 1248 | F>L | No |
TOPMed gnomAD |
|
| rs561557672 | 1251 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1273689981 | 1252 | R>C | No |
TOPMed gnomAD |
|
| rs201358234 | 1252 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1253 | V>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs377066371 | 1253 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1253 | V>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753108980 | 1253 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs753108980 | 1253 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1181614847 | 1254 | T>A | No |
TOPMed gnomAD |
|
| rs372464024 | 1254 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs372464024 | 1254 | T>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs912575096 | 1256 | Q>H | No |
TOPMed gnomAD |
|
| rs944048949 | 1256 | Q>R | No | TOPMed | |
| rs762086583 | 1257 | L>F | No |
ExAC gnomAD |
|
| rs1424384992 | 1258 | S>G | No | gnomAD | |
| rs12678871 | 1258 | S>I | No | Ensembl | |
| rs1407840797 | 1259 | H>Q | No |
TOPMed gnomAD |
|
|
rs768756951 TCGA novel |
1263 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC TOPMed gnomAD |
| rs1809625054 | 1263 | M>T | No | TOPMed | |
| rs768756951 | 1263 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs747553029 | 1264 | Q>E | No |
ExAC gnomAD |
|
| rs372689337 | 1264 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs375451644 | 1268 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM3899472 COSM3899471 rs374014840 |
1268 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| COSM275728 | 1270 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753509697 | 1272 | C>F | No |
1000Genomes ExAC gnomAD |
|
| rs753509697 | 1272 | C>S | No |
1000Genomes ExAC gnomAD |
|
| rs36068949 | 1274 | N>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV000975184 rs36068949 |
1274 | N>S | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1586800471 | 1276 | H>P | No | Ensembl | |
| rs375966959 | 1276 | H>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs755648168 | 1276 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs371595587 | 1277 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs371595587 | 1277 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs368694527 | 1278 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs766179392 | 1278 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs761987618 | 1279 | Q>* | No |
ExAC gnomAD |
|
| rs2129671961 | 1279 | Q>L | No | Ensembl | |
| rs760747840 | 1280 | G>S | No |
ExAC TOPMed gnomAD |
|
|
COSM1098968 COSM1098970 rs775604707 |
1281 | F>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2133623066 | 1282 | A>V | No | Ensembl | |
| rs767563825 | 1283 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1218895759 | 1283 | Q>H | No | gnomAD | |
| TCGA novel | 1285 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs564745975 | 1286 | L>P | No |
1000Genomes ExAC gnomAD |
|
| rs760077366 | 1286 | L>V | No |
ExAC gnomAD |
|
| TCGA novel | 1287 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1225259268 | 1287 | K>I | No | gnomAD | |
| rs1287634008 | 1288 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1348491048 | 1288 | K>R | No | gnomAD | |
| rs372878950 | 1289 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1809452584 | 1291 | H>L | No |
TOPMed gnomAD |
|
| rs778014893 | 1292 | R>* | No | gnomAD | |
| rs778014893 | 1292 | R>G | No | gnomAD | |
|
COSM2788916 rs749611889 COSM2788917 |
1292 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1809451790 | 1295 | I>V | No | TOPMed | |
| rs1586795835 | 1296 | P>H | No | Ensembl | |
| rs769633398 | 1296 | P>S | No |
ExAC gnomAD |
|
| rs2133622822 | 1297 | G>D | No | Ensembl | |
| rs1160221115 | 1297 | G>S | No | gnomAD | |
| rs747784923 | 1298 | C>F | No |
ExAC gnomAD |
|
| rs1196567562 | 1301 | T>I | No |
TOPMed gnomAD |
|
| rs1196567562 | 1301 | T>N | No |
TOPMed gnomAD |
|
| rs74469345 | 1303 | E>* | No | Ensembl | |
| rs780894636 | 1306 | S>C | No |
ExAC gnomAD |
|
| rs754504315 | 1307 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1270349457 | 1308 | I>T | No | gnomAD | |
| rs1193388559 | 1309 | P>S | No | Ensembl | |
| rs1455947049 | 1310 | E>D | No |
TOPMed gnomAD |
|
| rs556198096 | 1310 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1310260663 | 1311 | D>G | No |
TOPMed gnomAD |
|
| rs1308663256 | 1311 | D>H | No | gnomAD | |
| rs1254164152 | 1312 | A>V | No | gnomAD | |
| rs773680900 | 1313 | Q>* | No |
ExAC gnomAD |
|
| rs1009724933 | 1313 | Q>R | No |
TOPMed gnomAD |
|
| rs770257103 | 1317 | E>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs770257103 | 1317 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1462550226 | 1318 | R>Q | No |
TOPMed gnomAD |
|
| rs576889281 | 1318 | R>W | No |
1000Genomes ExAC gnomAD |
|
|
COSM261657 rs553766326 |
1320 | A>S | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
| rs553766326 | 1320 | A>T | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 1323 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1480989185 | 1324 | M>I | No |
TOPMed gnomAD |
|
| rs779860474 | 1324 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1301600622 | 1325 | A>P | No | gnomAD | |
|
COSM1496825 COSM1496826 rs1301600622 |
1325 | A>T | kidney [Cosmic] | No |
cosmic curated gnomAD |
|
rs772114436 COSM3899470 COSM3899469 |
1326 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs745771696 | 1327 | N>S | No |
ExAC gnomAD |
|
| rs933877920 | 1328 | V>A | No | TOPMed | |
| rs1433029018 | 1331 | P>L | No |
TOPMed gnomAD |
|
| rs1809280669 | 1332 | A>T | No | TOPMed | |
| rs752941943 | 1336 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs752941943 | 1336 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs2133610877 | 1337 | E>G | No | Ensembl | |
| rs1809280107 | 1337 | E>Q | No | TOPMed | |
| rs755020170 | 1338 | A>G | No |
ExAC gnomAD |
|
| rs1446225834 | 1338 | A>P | No |
TOPMed gnomAD |
|
| rs1167942266 | 1339 | Y>C | No | TOPMed | |
| rs1480933525 | 1340 | I>T | No | gnomAD | |
| TCGA novel | 1342 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1098965 COSM1098967 |
1344 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763561548 | 1345 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs750926603 | 1346 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1365031170 | 1346 | S>N | No | Ensembl | |
| rs185621129 | 1346 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1311090326 | 1347 | V>A | No | gnomAD | |
| rs201916564 | 1347 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1809277627 | 1348 | L>P | No | TOPMed | |
| rs1809277463 | 1349 | A>T | No | Ensembl | |
| rs1395919144 | 1351 | E>Q | No | gnomAD | |
| rs1809277199 | 1351 | E>V | No | Ensembl | |
| rs975439251 | 1352 | N>Y | No | TOPMed | |
| rs1397841348 | 1353 | L>F | No | gnomAD | |
| rs1427246929 | 1355 | T>S | No | gnomAD | |
| rs1369476648 | 1356 | L>S | No |
TOPMed gnomAD |
|
| rs1809276282 | 1357 | D>E | No | TOPMed | |
| rs768287004 | 1357 | D>Y | No |
ExAC gnomAD |
|
|
COSM3648099 COSM3648098 rs760240202 |
1358 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs760240202 | 1358 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM3899468 COSM3899467 rs775114146 |
1358 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs771451545 | 1359 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1253902563 | 1360 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
rs745847947 COSM5642422 COSM5642421 |
1360 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1274061636 | 1361 | Q>P | No | gnomAD | |
|
COSM3432329 COSM3432330 rs754322224 |
1364 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1194722507 | 1364 | A>V | No | TOPMed | |
| rs1437997799 | 1365 | V>A | No |
TOPMed gnomAD |
|
| rs1437997799 | 1365 | V>G | No |
TOPMed gnomAD |
|
| rs1809253144 | 1371 | G>R | No | gnomAD | |
| rs764353239 | 1372 | K>E | No |
ExAC gnomAD |
|
| rs1322464349 | 1373 | G>R | No | gnomAD | |
| rs760415108 | 1374 | K>R | No |
ExAC gnomAD |
|
| rs377382162 | 1376 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1057211649 | 1378 | R>S | No |
TOPMed gnomAD |
|
| rs758987621 | 1381 | S>G | No |
ExAC gnomAD |
|
| rs1809251387 | 1382 | S>F | No | Ensembl | |
| TCGA novel | 1385 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1376037682 | 1386 | N>S | No | gnomAD | |
| rs1238451502 | 1388 | L>F | No | gnomAD | |
| rs767009385 | 1389 | S>C | No |
ExAC gnomAD |
|
| rs768083592 | 1392 | R>* | No |
TOPMed gnomAD |
|
| rs759181929 | 1392 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs773904580 | 1393 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs990730752 | 1393 | Q>R | No |
TOPMed gnomAD |
|
| rs1809183247 | 1394 | D>H | No | Ensembl | |
| TCGA novel | 1394 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765849334 | 1395 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs765849334 | 1395 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1285758574 | 1396 | I>V | No | gnomAD | |
| rs1445681991 | 1397 | P>S | No | gnomAD | |
| rs906860624 | 1398 | S>L | No |
TOPMed gnomAD |
|
| rs1809182169 | 1398 | S>P | No | TOPMed | |
| rs747841646 | 1399 | Y>* | No |
ExAC gnomAD |
|
| rs1809181698 | 1399 | Y>C | No | Ensembl | |
| rs769656377 | 1399 | Y>H | No |
ExAC gnomAD |
|
| rs143504824 | 1400 | S>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs143504824 | 1400 | S>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1021017753 | 1402 | G>S | No |
TOPMed gnomAD |
|
| rs374297067 | 1403 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1380189517 | 1404 | N>I | No |
TOPMed gnomAD |
|
| rs1380189517 | 1404 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs368888774 | 1405 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1230098851 | 1406 | G>S | No | gnomAD | |
| rs749258265 | 1407 | R>L | No |
ExAC gnomAD |
|
| rs749258265 | 1407 | R>Q | No |
ExAC gnomAD |
|
| rs368179606 | 1407 | R>W | No |
ESP TOPMed gnomAD |
|
| rs1434645692 | 1412 | Q>H | No | gnomAD | |
| rs1295475018 | 1412 | Q>R | No |
TOPMed gnomAD |
|
| rs1190664692 | 1413 | D>A | No | gnomAD | |
| rs1190664692 | 1413 | D>V | No | gnomAD | |
| rs1808679349 | 1415 | S>F | No |
TOPMed gnomAD |
|
|
COSM3648096 COSM3648097 |
1415 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1808679228 | 1417 | T>I | No | gnomAD | |
| rs1354771377 | 1418 | T>I | No | gnomAD | |
| rs1415285417 | 1419 | V>I | No | TOPMed | |
| rs748662647 | 1421 | R>K | No |
ExAC gnomAD |
|
| rs1808678202 | 1422 | G>V | No | TOPMed | |
| rs1586773737 | 1423 | I>L | No | Ensembl | |
| rs781572213 | 1423 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1586773737 | 1423 | I>V | No | Ensembl | |
| rs1433256877 | 1424 | A>D | No |
TOPMed gnomAD |
|
| rs1159154849 | 1424 | A>P | No |
TOPMed gnomAD |
|
| rs1159154849 | 1424 | A>S | No |
TOPMed gnomAD |
|
| rs1433256877 | 1424 | A>V | No |
TOPMed gnomAD |
|
| rs1210991036 | 1425 | P>S | No |
TOPMed gnomAD |
|
| rs374575291 | 1426 | A>S | No |
ESP TOPMed gnomAD |
|
| rs1808676860 | 1426 | A>V | No | gnomAD | |
| rs201933345 | 1427 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs749969830 | 1428 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs373668415 | 1430 | S>C | No |
ESP ExAC gnomAD |
|
| rs1018387226 | 1431 | V>L | No | Ensembl | |
| rs753777836 | 1433 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs991270539 | 1433 | P>S | No |
TOPMed gnomAD |
|
| rs764002928 | 1434 | Q>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1435 | N>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774718387 | 1437 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs760487348 | 1437 | H>Y | No |
ExAC gnomAD |
|
| rs1808674453 | 1438 | S>Y | No | Ensembl | |
| rs771344374 | 1439 | P>L | No |
ExAC gnomAD |
|
| rs771344374 | 1439 | P>R | No |
ExAC gnomAD |
|
| rs1437704475 | 1441 | P>T | No | gnomAD | |
| rs773440014 | 1442 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs541841891 | 1442 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs773440014 | 1442 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs765258742 | 1443 | L>F | No |
ExAC gnomAD |
|
| rs765258742 | 1443 | L>I | No |
ExAC gnomAD |
|
| rs1048830108 | 1445 | N>K | No | TOPMed | |
| rs1166806892 | 1446 | L>H | No | TOPMed | |
| rs369865951 | 1446 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1253979482 | 1447 | A>G | No | gnomAD | |
| rs1808365785 | 1447 | A>S | No | TOPMed | |
| rs1253979482 | 1447 | A>V | No | gnomAD | |
| rs929092774 | 1448 | A>S | No |
TOPMed gnomAD |
|
| rs769220820 | 1448 | A>V | No |
ExAC gnomAD |
|
| rs1207504620 | 1450 | Y>S | No | gnomAD | |
| rs377446510 | 1453 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1808364644 | 1453 | P>L | No | Ensembl | |
| rs377446510 | 1453 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs377446510 | 1453 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1246004618 | 1455 | K>R | No |
TOPMed gnomAD |
|
| rs1808363956 | 1456 | S>F | No | TOPMed | |
| rs1323941990 | 1456 | S>T | No | gnomAD | |
| rs775884382 | 1457 | F>L | No |
ExAC gnomAD |
|
| TCGA novel | 1457 | F>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200599694 | 1458 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200599694 | 1458 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1563693172 | 1459 | P>L | No | TOPMed | |
| rs1586764761 | 1461 | M>T | No | Ensembl | |
| rs1808363044 | 1462 | P>Q | No | Ensembl | |
| rs756724342 | 1463 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs748872453 | 1463 | K>M | No |
ExAC gnomAD |
|
| rs1326952267 | 1464 | L>I | No |
TOPMed gnomAD |
|
| rs777406920 | 1466 | K>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1468 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752648287 | 1469 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1306400645 | 1470 | P>L | No | Ensembl | |
| rs1472892267 | 1470 | P>S | No | gnomAD | |
|
VAR_055982 rs17526980 |
1471 | V>I | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs776750868 | 1472 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs776750868 | 1472 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs764139083 | 1472 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs764139083 | 1472 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1448046330 | 1473 | D>E | No | Ensembl | |
| rs772336597 | 1473 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs201563049 | 1473 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1289615085 | 1474 | E>D | No | gnomAD | |
| rs1330432646 | 1474 | E>K | No | gnomAD | |
| rs2133549899 | 1475 | K>E | No | Ensembl | |
| rs1808359390 | 1476 | R>G | No | TOPMed | |
| rs1226022474 | 1476 | R>S | No | gnomAD | |
| rs1323769241 | 1477 | G>D | No | gnomAD | |
| rs746177089 | 1477 | G>S | No |
ExAC gnomAD |
|
| rs915321754 | 1478 | K>E | No | Ensembl | |
| rs774572916 | 1479 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs770673480 | 1479 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs774572916 | 1479 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs201077981 | 1480 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201077981 | 1480 | P>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201077981 | 1480 | P>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781130442 | 1482 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1230203216 | 1483 | L>P | No |
TOPMed gnomAD |
|
| rs561619897 | 1484 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1360712029 | 1485 | H>R | No | Ensembl | |
| rs779703300 | 1485 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1807506092 | 1487 | P>S | No |
TOPMed gnomAD |
|
| rs1807506092 | 1487 | P>T | No |
TOPMed gnomAD |
|
| rs199652125 | 1488 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1807505367 | 1489 | P>R | No | gnomAD | |
| rs1225474229 | 1489 | P>S | No |
TOPMed gnomAD |
|
| rs1225474229 | 1489 | P>T | No |
TOPMed gnomAD |
|
| rs201273629 | 1490 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs767471422 COSM5976814 COSM5976813 |
1490 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1166527015 | 1492 | M>I | No |
TOPMed gnomAD |
|
| rs1807504637 | 1493 | V>M | No | Ensembl | |
| rs867385353 | 1494 | Q>* | No | gnomAD | |
| rs760126656 | 1494 | Q>R | No |
ExAC gnomAD |
|
| rs1277290870 | 1495 | S>P | No | gnomAD | |
| rs1275302709 | 1496 | A>V | No | Ensembl | |
| rs1484387808 | 1497 | S>G | No |
TOPMed gnomAD |
|
| rs1256251747 | 1498 | P>A | No | gnomAD | |
|
rs374209248 COSM1456522 COSM1456523 |
1498 | P>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs374209248 | 1498 | P>Q | No |
TOPMed gnomAD |
|
| rs374209248 | 1498 | P>R | No |
TOPMed gnomAD |
|
| rs1256251747 | 1498 | P>S | No | gnomAD | |
| TCGA novel | 1499 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs368364212 | 1500 | I>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs368364212 | 1500 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs867241022 | 1501 | R>G | No | TOPMed | |
| rs868361626 | 1501 | R>M | No | Ensembl | |
| rs200434029 | 1503 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs200434029 | 1503 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs200434029 | 1503 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs371444377 | 1504 | R>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1353586484 | 1505 | M>I | No | gnomAD | |
| rs776257053 | 1505 | M>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1507 | E>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2133481036 | 1507 | E>D | No | Ensembl | |
| rs2133469090 | 1508 | A>D | No | Ensembl | |
| rs772053981 | 1509 | Q>* | No |
ExAC gnomAD |
|
| rs1308638938 | 1510 | P>A | No | TOPMed | |
| rs770317379 | 1510 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs770317379 | 1510 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs774171526 | 1511 | E>A | No |
ExAC gnomAD |
|
| rs202077289 | 1512 | M>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs369153052 | 1513 | G>D | No |
ESP TOPMed |
|
| rs1807335384 | 1514 | P>R | No | TOPMed | |
| rs1586737956 | 1516 | V>A | No | Ensembl | |
| rs1023818789 | 1516 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs879120984 | 1518 | V>L | No |
TOPMed gnomAD |
|
| rs879120984 | 1518 | V>M | No |
TOPMed gnomAD |
|
| rs1807333914 | 1519 | Q>K | No | Ensembl | |
| rs1305971481 | 1519 | Q>R | No | gnomAD | |
| rs1439152681 | 1520 | T>K | No |
TOPMed gnomAD |
|
| rs1443590999 | 1521 | M>I | No | gnomAD | |
| rs755086480 | 1521 | M>V | No |
ExAC gnomAD |
|
| rs1807332887 | 1522 | G>A | No | TOPMed | |
| rs1342886622 | 1523 | A>G | No | gnomAD | |
| rs1433031821 | 1523 | A>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1455823629 | 1523 | A>S | No | gnomAD | |
| rs1157734865 | 1524 | P>L | No |
TOPMed gnomAD |
|
| rs1785566813 | 1524 | P>S | No | TOPMed | |
| rs1033074771 | 1527 | K>N | No | TOPMed | |
| rs775030095 | 1528 | I>M | No |
ExAC gnomAD |
|
| rs369401468 | 1528 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1807331422 | 1529 | C>Y | No | TOPMed | |
| rs900113829 | 1530 | D>G | No | Ensembl | |
| rs750932932 | 1530 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs750932932 | 1530 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs750932932 | 1530 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1586737815 | 1531 | K>N | No | Ensembl | |
| rs757515152 | 1533 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs1807330626 | 1533 | A>T | No | Ensembl | |
| rs757515152 | 1533 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1484870909 | 1534 | K>E | No |
TOPMed gnomAD |
|
| rs1484870909 | 1534 | K>Q | No |
TOPMed gnomAD |
|
| rs1050903600 | 1536 | P>A | No |
TOPMed gnomAD |
|
| rs753544473 | 1536 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1050903600 | 1536 | P>S | No |
TOPMed gnomAD |
|
| rs1050903600 | 1536 | P>T | No |
TOPMed gnomAD |
|
| rs1807329229 | 1537 | S>C | No |
TOPMed gnomAD |
|
| rs1807329229 | 1537 | S>F | No |
TOPMed gnomAD |
|
| rs200102612 | 1537 | S>P | No | Ensembl | |
| rs117202308 | 1539 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1232431017 | 1540 | P>S | No | gnomAD | |
| rs1226131203 | 1541 | V>D | No |
TOPMed gnomAD |
|
| rs766983278 | 1542 | I>R | No |
ExAC TOPMed gnomAD |
|
| rs766983278 | 1542 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1807327978 | 1543 | A>T | No | TOPMed | |
| rs759500649 | 1545 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs774220954 | 1546 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1360505133 | 1548 | T>S | No |
TOPMed gnomAD |
|
| rs367772924 | 1549 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs367772924 | 1549 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs941257095 | 1551 | P>L | No |
TOPMed gnomAD |
|
| rs1807325679 | 1552 | E>G | No | TOPMed | |
| rs1171344593 | 1553 | A>V | No | gnomAD | |
| rs576137578 | 1554 | Q>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs747214816 | 1555 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1426607352 | 1556 | G>R | No |
TOPMed gnomAD |
|
| rs1163407445 | 1557 | P>L | No | gnomAD | |
|
COSM1456521 COSM1456520 rs1428654026 |
1557 | P>S | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1428654026 | 1557 | P>T | No |
TOPMed gnomAD |
|
| rs1807324037 | 1558 | P>S | No | TOPMed | |
| TCGA novel | 1559 | S>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1807323657 | 1559 | S>N | No |
TOPMed gnomAD |
|
| rs780325964 | 1559 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs772239580 | 1560 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs772239580 | 1560 | P>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1561 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1275721543 | 1565 | S>F | No |
TOPMed gnomAD |
|
| rs1586737552 | 1569 | F>S | No | Ensembl | |
| rs75022514 | 1570 | S>P | No | Ensembl | |
| rs1356761459 | 1571 | H>R | No | gnomAD | |
| rs1182061064 | 1571 | H>Y | No |
TOPMed gnomAD |
|
| rs1807321277 | 1573 | V>I | No | TOPMed | |
| rs767032667 | 1575 | T>I | No |
ExAC gnomAD |
|
| rs751500202 | 1576 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs766290711 | 1576 | A>V | No |
ExAC gnomAD |
|
| rs769605242 | 1579 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1807319956 | 1580 | D>N | No | TOPMed | |
| rs1420025061 | 1581 | A>S | No |
TOPMed gnomAD |
|
| rs1420025061 | 1581 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1807319468 | 1582 | L>P | No | Ensembl | |
| rs1464964973 | 1583 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1807317510 | 1584 | P>H | No | Ensembl | |
| rs2133467805 | 1585 | G>S | No | Ensembl | |
| rs916728768 | 1589 | A>G | No | TOPMed | |
| rs1788013424 | 1589 | A>T | No | Ensembl | |
| rs1807316112 | 1591 | P>L | No | TOPMed | |
| rs1807315682 | 1593 | G>E | No | TOPMed | |
| rs1251132177 | 1594 | S>C | No |
TOPMed gnomAD |
|
| rs1251132177 | 1594 | S>F | No |
TOPMed gnomAD |
|
| rs745879040 | 1595 | M>V | No |
ExAC TOPMed |
|
| rs576809142 | 1596 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs576809142 | 1596 | P>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1286461756 | 1597 | T>I | No | Ensembl | |
| rs1286461756 | 1597 | T>N | No | Ensembl | |
| rs1031287048 | 1598 | A>V | No | gnomAD | |
| rs1327333684 | 1599 | P>A | No |
TOPMed gnomAD |
|
| rs553708749 | 1600 | E>G | No |
1000Genomes gnomAD |
|
| rs1227944519 | 1601 | A>T | No |
1000Genomes gnomAD |
|
| rs1807313813 | 1601 | A>V | No | TOPMed | |
| rs1807313561 | 1602 | E>K | No | TOPMed | |
| rs1807313282 | 1603 | P>H | No | Ensembl | |
| rs1807313425 | 1603 | P>S | No | Ensembl | |
| rs2133467422 | 1604 | E>K | No | Ensembl | |
| rs1807312831 | 1605 | A>V | No |
TOPMed gnomAD |
|
| rs1807312538 | 1606 | P>S | No | gnomAD | |
| rs1807312254 | 1608 | S>C | No | 1000Genomes | |
| rs974080087 | 1609 | H>P | No | Ensembl | |
| rs1586737306 | 1609 | H>Y | No | Ensembl | |
| rs866631138 | 1610 | P>L | No | Ensembl | |
| rs1807311695 | 1610 | P>S | No | TOPMed | |
| rs1807311136 | 1611 | P>Q | No | Ensembl | |
| rs1422979172 | 1611 | P>S | No |
TOPMed gnomAD |
|
| rs1422979172 | 1611 | P>T | No |
TOPMed gnomAD |
|
| rs1807310835 | 1612 | P>L | No | gnomAD | |
| rs1807310835 | 1612 | P>Q | No | gnomAD | |
| rs1807310448 | 1614 | T>M | No | Ensembl | |
| rs1232139228 | 1615 | A>T | No | gnomAD | |
| rs1807310098 | 1615 | A>V | No |
TOPMed gnomAD |
|
| rs1367478541 | 1616 | V>A | No | gnomAD | |
| rs1471414893 | 1616 | V>I | No | TOPMed | |
| rs1586737223 | 1617 | P>R | No | Ensembl | |
| rs1436454561 | 1618 | A>T | No |
TOPMed gnomAD |
|
| rs1173922237 | 1619 | E>K | No |
TOPMed gnomAD |
|
| rs1350613947 | 1621 | P>A | No |
1000Genomes TOPMed gnomAD |
|
| rs1350613947 | 1621 | P>T | No |
1000Genomes TOPMed gnomAD |
|
| rs1406724373 | 1622 | P>R | No |
TOPMed gnomAD |
|
| rs1387870263 | 1623 | G>S | No |
TOPMed gnomAD |
|
| rs1807308043 | 1623 | G>V | No | TOPMed | |
| rs899609149 | 1624 | P>L | No |
TOPMed gnomAD |
|
| rs899609149 | 1624 | P>R | No |
TOPMed gnomAD |
|
| rs1807307920 | 1624 | P>S | No | TOPMed | |
| rs534256791 | 1626 | Q>H | No |
1000Genomes TOPMed gnomAD |
|
| rs1293897813 | 1627 | L>F | No |
TOPMed gnomAD |
|
| rs1462893512 | 1628 | V>M | No |
TOPMed gnomAD |
|
| rs1349245798 | 1629 | S>N | No |
TOPMed gnomAD |
|
| rs1392181197 | 1629 | S>R | No | gnomAD | |
| rs1349245798 | 1629 | S>T | No |
TOPMed gnomAD |
|
| rs1213577317 | 1631 | G>A | No |
TOPMed gnomAD |
|
| rs1164204209 | 1631 | G>S | No | gnomAD | |
| rs1474905484 | 1632 | R>Q | No |
TOPMed gnomAD |
|
| rs1249374620 | 1632 | R>W | No |
TOPMed gnomAD |
|
| rs1005368278 | 1633 | E>G | No | gnomAD | |
| rs1411199048 | 1634 | R>C | No | TOPMed | |
| rs1807305672 | 1634 | R>H | No |
TOPMed gnomAD |
|
| rs1807305672 | 1634 | R>L | No |
TOPMed gnomAD |
|
| rs1807305534 | 1635 | P>S | No | Ensembl | |
| rs574810703 | 1636 | D>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs756451345 | 1636 | D>N | No |
ExAC gnomAD |
|
| rs888352370 | 1637 | L>F | No |
TOPMed gnomAD |
|
| rs1210825080 | 1638 | E>K | No | gnomAD | |
| rs1470156567 | 1640 | P>L | No | gnomAD | |
| rs1807304007 | 1640 | P>S | No | TOPMed | |
| rs1213672375 | 1641 | A>T | No |
TOPMed gnomAD |
|
| rs1329181913 | 1641 | A>V | No |
TOPMed gnomAD |
|
| rs1262007130 | 1642 | P>R | No |
TOPMed gnomAD |
|
| rs1236344519 | 1643 | G>C | No | gnomAD | |
| rs1236344519 | 1643 | G>S | No | gnomAD | |
| rs554817518 | 1644 | S>Y | No |
1000Genomes ExAC gnomAD |
|
| rs1329860970 | 1645 | P>L | No | TOPMed | |
| rs1329860970 | 1645 | P>Q | No | TOPMed | |
| rs1329860970 | 1645 | P>R | No | TOPMed | |
| rs1442820468 | 1645 | P>S | No | gnomAD | |
| rs2133466445 | 1646 | F>L | No | 1000Genomes | |
| rs754617977 | 1647 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs754617977 | 1647 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1807301742 | 1647 | R>H | No | Ensembl | |
| rs751124305 | 1648 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs13263056 | 1649 | R>L | No | TOPMed | |
| rs13263056 | 1649 | R>Q | No | TOPMed | |
| rs1160378572 | 1649 | R>W | No |
TOPMed gnomAD |
|
| rs1807300800 | 1650 | R>M | No |
TOPMed gnomAD |
|
| rs1305565617 | 1651 | V>A | No | gnomAD | |
| rs1363427123 | 1651 | V>L | No | gnomAD | |
| rs1363427123 | 1651 | V>M | No | gnomAD | |
| rs1807299962 | 1652 | R>Q | No | Ensembl | |
| rs1194575791 | 1652 | R>W | No | gnomAD | |
| rs1237189431 | 1653 | A>T | No |
TOPMed gnomAD |
|
| rs1455836285 | 1654 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1455836285 | 1654 | S>W | No |
TOPMed gnomAD |
|
| rs1339511787 | 1655 | E>Q | No |
TOPMed gnomAD |
|
| rs1193515874 | 1657 | R>C | No |
TOPMed gnomAD |
|
| rs1193515874 | 1657 | R>G | No |
TOPMed gnomAD |
|
| rs1448086506 | 1657 | R>H | No |
TOPMed gnomAD |
|
| rs1448086506 | 1657 | R>L | No |
TOPMed gnomAD |
|
| rs1448086506 | 1657 | R>P | No |
TOPMed gnomAD |
|
| rs1273354529 | 1658 | S>C | No | TOPMed | |
| rs538052005 | 1659 | F>S | No |
1000Genomes ExAC gnomAD |
|
| rs538052005 | 1659 | F>Y | No |
1000Genomes ExAC gnomAD |
|
| rs1218848641 | 1660 | S>L | No |
TOPMed gnomAD |
|
| rs755491671 | 1661 | R>C | No |
TOPMed gnomAD |
|
| rs1345413811 | 1661 | R>H | No | gnomAD | |
| rs1345413811 | 1661 | R>L | No | gnomAD | |
| rs755491671 | 1661 | R>S | No |
TOPMed gnomAD |
|
| rs772358686 | 1662 | M>I | No |
TOPMed gnomAD |
|
| rs1302456071 | 1662 | M>L | No |
TOPMed gnomAD |
|
| rs569394326 | 1662 | M>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1302456071 | 1662 | M>V | No |
TOPMed gnomAD |
|
| rs1328143260 | 1664 | A>S | No |
TOPMed gnomAD |
|
| rs1328143260 | 1664 | A>T | No |
TOPMed gnomAD |
|
| rs1316210351 | 1664 | A>V | No |
TOPMed gnomAD |
|
| rs764968047 | 1665 | G>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1665 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1386698072 | 1667 | P>A | No | gnomAD | |
| rs1436848926 | 1668 | G>S | No |
TOPMed gnomAD |
|
| rs917253629 | 1670 | S>F | No |
TOPMed gnomAD |
|
| rs1807295820 | 1670 | S>P | No | TOPMed | |
| rs917253629 | 1670 | S>Y | No |
TOPMed gnomAD |
|
| rs989640527 | 1671 | P>L | No |
TOPMed gnomAD |
|
| rs989640527 | 1671 | P>R | No |
TOPMed gnomAD |
|
| rs1173415987 | 1671 | P>S | No | gnomAD | |
| rs1191713559 | 1672 | G>W | No | gnomAD | |
| rs1257633306 | 1673 | A>D | No | gnomAD | |
| rs1257633306 | 1673 | A>G | No | gnomAD | |
| rs761642628 | 1673 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs761642628 | 1673 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs775861530 | 1674 | E>D | No |
ExAC gnomAD |
|
| rs978333948 | 1674 | E>K | No | TOPMed | |
| rs1306976598 | 1675 | G>V | No | gnomAD | |
| rs992882593 | 1675 | G>W | No |
TOPMed gnomAD |
|
| rs1214927565 | 1677 | A>P | No |
TOPMed gnomAD |
|
| rs1214927565 | 1677 | A>S | No |
TOPMed gnomAD |
|
| rs1214927565 | 1677 | A>T | No |
TOPMed gnomAD |
|
| rs1341296675 | 1677 | A>V | No |
TOPMed gnomAD |
|
| rs748528379 | 1678 | P>A | No |
TOPMed gnomAD |
|
| rs1226630898 | 1678 | P>L | No |
TOPMed gnomAD |
|
| rs748528379 | 1678 | P>S | No |
TOPMed gnomAD |
|
| rs1416791217 | 1679 | A>G | No |
TOPMed gnomAD |
|
| rs1313563583 | 1679 | A>T | No | gnomAD | |
| rs1416791217 | 1679 | A>V | No |
TOPMed gnomAD |
|
| rs1312569027 | 1680 | P>L | No | gnomAD | |
| rs1373850491 | 1680 | P>T | No | gnomAD | |
| rs1807291198 | 1681 | G>C | No | Ensembl | |
| rs1807290927 | 1681 | G>V | No | Ensembl | |
| rs1807290602 | 1682 | A>P | No | TOPMed | |
| rs1807290602 | 1682 | A>T | No | TOPMed | |
| rs772491346 | 1682 | A>V | No |
ExAC TOPMed gnomAD |
|
|
rs981072011 TCGA novel |
1683 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs981072011 | 1683 | G>W | No |
TOPMed gnomAD |
|
| rs1189467589 | 1684 | G>E | No | gnomAD | |
| rs1417872356 | 1684 | G>R | No |
TOPMed gnomAD |
|
| rs1484438426 | 1685 | Q>K | No | gnomAD | |
| rs1204828151 | 1687 | L>P | No |
TOPMed gnomAD |
|
| rs1204828151 | 1687 | L>R | No |
TOPMed gnomAD |
|
| rs1320802581 | 1688 | A>V | No |
TOPMed gnomAD |
|
| rs1484197514 | 1689 | S>P | No |
TOPMed gnomAD |
|
| rs771002478 | 1691 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs1335406484 | 1692 | E>K | No |
TOPMed gnomAD |
|
| rs1022102959 | 1694 | A>D | No |
TOPMed gnomAD |
|
| rs1022102959 | 1694 | A>V | No |
TOPMed gnomAD |
|
| rs749780737 | 1695 | D>E | No |
ExAC gnomAD |
|
| rs1807287690 | 1696 | E>D | No | Ensembl | |
| rs1355703270 | 1697 | V>A | No |
TOPMed gnomAD |
|
| rs1586736508 | 1697 | V>I | No | Ensembl | |
| rs1009322807 | 1698 | P>L | No | gnomAD | |
| rs1009322807 | 1698 | P>R | No | gnomAD | |
| rs1356605252 | 1699 | E>A | No |
TOPMed gnomAD |
|
| rs566786202 | 1699 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 1700 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1205691203 | 1702 | R>* | No | Ensembl | |
| rs1807286210 | 1704 | G>S | No | TOPMed | |
| rs1409842586 | 1704 | G>V | No | gnomAD | |
| TCGA novel | 1705 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs367988811 | 1706 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1456518 COSM1456519 rs1473922547 |
1707 | V>I | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1473922547 | 1707 | V>L | No |
TOPMed gnomAD |
|
| rs375844229 | 1708 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs375844229 | 1708 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1230906498 | 1709 | V>L | No |
TOPMed gnomAD |
|
| rs1230906498 | 1709 | V>M | No |
TOPMed gnomAD |
|
| rs757880042 | 1711 | A>P | No |
ExAC gnomAD |
|
| rs757880042 | 1711 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs941581834 | 1712 | H>Q | No |
TOPMed gnomAD |
|
| rs1039976250 | 1712 | H>R | No | Ensembl | |
| rs547030704 | 1714 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs547030704 | 1714 | T>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs779345859 | 1716 | V>L | No |
ExAC TOPMed gnomAD |
|
|
COSM1212357 COSM1212356 rs779345859 |
1716 | V>M | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1375820012 | 1718 | R>T | No | gnomAD | |
| rs1025914110 | 1719 | Y>* | No |
1000Genomes TOPMed gnomAD |
|
|
rs200418743 RCV000884441 |
1719 | Y>C | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs753746284 | 1720 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs753746284 | 1720 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs917366507 | 1723 | A>S | No |
TOPMed gnomAD |
|
| TCGA novel | 1723 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763949498 | 1723 | A>V | No | ExAC | |
| rs1159213137 | 1724 | D>H | No |
TOPMed gnomAD |
|
| rs1159213137 | 1724 | D>N | No |
TOPMed gnomAD |
|
| rs1401998179 | 1725 | F>L | No |
TOPMed gnomAD |
|
| rs1176305078 | 1727 | E>* | No |
TOPMed gnomAD |
|
| rs1176305078 | 1727 | E>K | No |
TOPMed gnomAD |
|
|
TCGA novel rs1807281343 |
1728 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs759867597 | 1728 | G>S | No |
ExAC gnomAD |
|
| rs2133464611 | 1729 | T>M | No | Ensembl | |
| rs1807280914 | 1730 | W>* | No | TOPMed | |
| rs1246146756 | 1732 | G>S | No | gnomAD | |
| rs1209996212 | 1736 | D>H | No |
TOPMed gnomAD |
|
| rs1209996212 | 1736 | D>N | No |
TOPMed gnomAD |
|
| rs1807279445 | 1738 | P>L | No | TOPMed | |
| rs1807279569 | 1738 | P>S | No | gnomAD | |
| rs1439267809 | 1739 | S>L | No | gnomAD | |
| rs1041378756 | 1740 | G>D | No | Ensembl | |
| rs745488043 | 1743 | D>E | No |
ExAC gnomAD |
|
| rs202164524 | 1744 | G>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs202164524 | 1744 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs202164524 | 1744 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs759416176 | 1746 | I>T | No |
TOPMed gnomAD |
|
| rs1807227754 | 1746 | I>V | No | TOPMed | |
| rs755997303 | 1747 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs752592040 | 1747 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs755997303 | 1747 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs755997303 | 1747 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs758684417 | 1748 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs758684417 | 1748 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs574109297 | 1748 | G>R | No |
1000Genomes TOPMed gnomAD |
|
| rs574109297 | 1748 | G>W | No |
1000Genomes TOPMed gnomAD |
|
| rs1033955710 | 1749 | K>E | No | gnomAD | |
| rs1179308343 | 1750 | Q>H | No | gnomAD | |
| rs1807226190 | 1750 | Q>R | No | gnomAD | |
| rs1807225932 | 1751 | Y>C | No | Ensembl | |
| rs1807225656 | 1752 | F>Y | No | TOPMed | |
| rs1586734741 | 1753 | R>G | No | Ensembl | |
| rs1260366037 | 1753 | R>S | No |
TOPMed gnomAD |
|
| rs1002125074 | 1754 | C>G | No | Ensembl | |
| rs1477957078 | 1755 | N>K | No |
TOPMed gnomAD |
|
| rs1586734714 | 1755 | N>T | No | Ensembl | |
| rs765325902 | 1756 | P>R | No |
ExAC gnomAD |
|
| rs1807224670 | 1756 | P>S | No | gnomAD | |
| rs965980979 | 1757 | G>D | No | TOPMed | |
| rs1195346670 | 1757 | G>S | No |
TOPMed gnomAD |
|
| rs965980979 | 1757 | G>V | No | TOPMed | |
| rs1586734683 | 1758 | Y>C | No | Ensembl | |
| rs1586734683 | 1758 | Y>F | No | Ensembl | |
| rs1270764810 | 1759 | G>E | No | gnomAD | |
| rs1336237194 | 1759 | G>R | No |
TOPMed gnomAD |
|
| rs761826952 | 1760 | L>P | No |
ExAC gnomAD |
|
| rs1017567808 | 1762 | V>A | No |
1000Genomes TOPMed gnomAD |
|
| rs1308777903 | 1762 | V>I | No | gnomAD | |
| rs764652302 | 1764 | P>H | No |
ExAC gnomAD |
|
| rs369595297 | 1765 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1288385028 | 1766 | R>Q | No |
TOPMed gnomAD |
|
| rs537017741 | 1766 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1402121308 | 1767 | V>G | No | gnomAD | |
| rs759254377 | 1768 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs770569664 | 1768 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs770569664 | 1768 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs952182226 | 1769 | R>G | No |
TOPMed gnomAD |
|
| rs1807220653 | 1769 | R>K | No |
TOPMed gnomAD |
|
| rs1807220500 | 1770 | A>T | No |
TOPMed gnomAD |
|
| rs1424659283 | 1771 | T>A | No | gnomAD | |
| rs569156581 | 1771 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1424659283 | 1771 | T>P | No | gnomAD | |
| rs781075161 | 1772 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1807219522 | 1772 | G>D | No | TOPMed | |
| rs781075161 | 1772 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1028219626 | 1773 | P>A | No | TOPMed | |
| rs779044932 | 1775 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs779044932 | 1775 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs754683695 | 1775 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs764060371 | 1776 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs764060371 | 1776 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs757466920 | 1776 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs552142818 | 1777 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs753094507 | 1777 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs753094507 | 1777 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1322888415 | 1778 | S>N | No | gnomAD | |
| rs759912737 | 1779 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs766188778 | 1780 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs566940732 | 1781 | L>H | No |
ExAC TOPMed gnomAD |
|
| rs1418137627 | 1781 | L>I | No | gnomAD | |
| rs566940732 | 1781 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs769143965 | 1782 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs772886058 COSM1734149 COSM1734148 |
1782 | R>W | pancreas [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs966456021 | 1783 | L>P | No |
TOPMed gnomAD |
|
| rs1447025097 | 1784 | G>A | No | gnomAD | |
| rs1447025097 | 1784 | G>D | No | gnomAD | |
| rs1243607028 | 1785 | A>T | No |
TOPMed gnomAD |
|
| rs748141536 | 1785 | A>V | No |
ExAC gnomAD |
|
| rs1353879225 | 1786 | P>L | No |
TOPMed gnomAD |
|
| rs776377730 | 1786 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs776377730 | 1786 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs757524983 | 1787 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs757524983 | 1787 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs945270217 | 1788 | A>D | No |
TOPMed gnomAD |
|
| rs749483065 | 1789 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs201015626 | 1789 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs753252153 | 1790 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs377365368 | 1790 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs560613937 | 1791 | S>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs755524267 | 1791 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs192432763 | 1792 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1400539555 | 1793 | T>I | No |
TOPMed gnomAD |
|
| rs1400539555 | 1793 | T>N | No |
TOPMed gnomAD |
|
| rs903803757 | 1793 | T>P | No | Ensembl | |
| rs1400539555 | 1793 | T>S | No |
TOPMed gnomAD |
|
| rs1458595566 | 1794 | L>F | No |
TOPMed gnomAD |
|
| rs1458595566 | 1794 | L>V | No |
TOPMed gnomAD |
|
| rs772693486 | 1795 | S>L | No |
ExAC gnomAD |
|
| rs896969746 | 1796 | G>D | No |
TOPMed gnomAD |
|
| rs1807211197 | 1796 | G>S | No | Ensembl | |
| rs2133459311 | 1797 | S>F | No | Ensembl | |
| rs768534631 | 1798 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs370028815 | 1798 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs768534631 | 1798 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1807210233 | 1799 | T>A | No | Ensembl | |
| rs1300660537 | 1799 | T>N | No |
TOPMed gnomAD |
|
| rs1586734192 | 1800 | N>H | No | Ensembl | |
| rs1212639331 | 1800 | N>I | No | gnomAD | |
| rs775305974 | 1802 | A>D | No |
ExAC gnomAD |
|
| rs775305974 | 1802 | A>V | No |
ExAC gnomAD |
|
|
COSM3925137 rs773872653 COSM3925136 |
1803 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs773872653 | 1803 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs777789673 | 1804 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1807208727 | 1805 | T>I | No | gnomAD | |
| rs371612114 | 1807 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs371612114 | 1807 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs375105823 | 1807 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1807207689 | 1809 | A>D | No | Ensembl | |
| rs1410935022 | 1809 | A>T | No |
TOPMed gnomAD |
|
| TCGA novel | 1809 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs368696178 | 1810 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1417981157 | 1811 | A>T | No | gnomAD | |
| rs200053841 | 1812 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs761512938 | 1812 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs753341839 | 1813 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1807205932 | 1813 | R>K | No | Ensembl | |
| rs763567000 | 1814 | S>N | No |
ExAC gnomAD |
|
| rs760639554 | 1815 | H>R | No |
ExAC gnomAD |
|
| rs1256051327 | 1815 | H>Y | No | gnomAD | |
| rs775359435 | 1816 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1348849392 | 1817 | N>D | No |
TOPMed gnomAD |
|
| rs1807204908 | 1817 | N>S | No | TOPMed | |
| rs1273893668 | 1818 | P>A | No |
TOPMed gnomAD |
|
| rs1230181124 | 1818 | P>L | No | gnomAD | |
| TCGA novel | 1819 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1283891781 | 1820 | N>S | No | gnomAD | |
| rs565579653 | 1821 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201770958 | 1821 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1822 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1807203375 | 1823 | S>Y | No | TOPMed | |
| rs781160340 | 1824 | W>* | No |
ExAC TOPMed gnomAD |
|
| rs781160340 | 1824 | W>C | No |
ExAC TOPMed gnomAD |
|
| rs768805349 | 1825 | A>V | No |
ExAC gnomAD |
No associated diseases with Q9NQT8
8 regional properties for Q9NQT8
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Forkhead-associated (FHA) domain | 472 - 535 | IPR000253 |
| domain | CAP Gly-rich domain | 1703 - 1768 | IPR000938 |
| domain | Kinesin motor domain | 3 - 361 | IPR001752 |
| conserved_site | Kinesin motor domain, conserved site | 247 - 258 | IPR019821 |
| domain | Kinesin-like KIF1-type | 756 - 802 | IPR022140 |
| domain | Kinesin-like | 1003 - 1058 | IPR022164-1 |
| domain | Kinesin-like | 1216 - 1279 | IPR022164-2 |
| domain | Kinesin-associated | 390 - 469 | IPR032405 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47117 | STAR-RELATED LIPID TRANSFER PROTEIN 9 |
| PANTHER Subfamily | PTHR47117:SF5 | SUBFAMILY NOT NAMED |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| kinesin complex | Any complex that includes a dimer of molecules from the kinesin superfamily, a group of related proteins that contain an extended region of predicted alpha-helical coiled coil in the main chain that likely produces dimerization. The native complexes of several kinesin family members have also been shown to contain additional peptides, often designated light chains as all of the noncatalytic subunits that are currently known are smaller than the chain that contains the motor unit. Kinesin complexes generally possess a force-generating enzymatic activity, or motor, which converts the free energy of the gamma phosphate bond of ATP into mechanical work. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| 14-3-3 protein binding | Binding to a 14-3-3 protein. A 14-3-3 protein is any of a large family of approximately 30kDa acidic proteins which exist primarily as homo- and heterodimers within all eukaryotic cells, and have been implicated in the modulation of distinct biological processes by binding to specific phosphorylated sites on diverse target proteins, thereby forcing conformational changes or influencing interactions between their targets and other molecules. Each 14-3-3 protein sequence can be roughly divided into three sections |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| microtubule binding | Binding to a microtubule, a filament composed of tubulin monomers. |
| microtubule motor activity | A motor activity that generates movement along a microtubule, driven by ATP hydrolysis. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| cytoskeleton-dependent intracellular transport | The directed movement of substances along cytoskeletal fibers such as microfilaments or microtubules within a cell. |
| microtubule-based movement | A microtubule-based process that results in the movement of organelles, other microtubules, or other cellular components. Examples include motor-driven movement along microtubules and movement driven by polymerization or depolymerization of microtubules. |
| protein targeting | The process of targeting specific proteins to particular regions of the cell, typically membrane-bounded subcellular organelles. Usually requires an organelle specific protein sequence motif. |
| regulation of axonogenesis | Any process that modulates the frequency, rate or extent of axonogenesis, the generation of an axon, the long process of a neuron. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| T cell activation | The change in morphology and behavior of a mature or immature T cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P33176 | KIF5B | Kinesin-1 heavy chain | Homo sapiens (Human) | EV |
| O60282 | KIF5C | Kinesin heavy chain isoform 5C | Homo sapiens (Human) | EV |
| Q12840 | KIF5A | Kinesin heavy chain isoform 5A | Homo sapiens (Human) | EV |
| Q2M1P5 | KIF7 | Kinesin-like protein KIF7 | Homo sapiens (Human) | EV |
| Q86VH2 | KIF27 | Kinesin-like protein KIF27 | Homo sapiens (Human) | SS |
| Q7Z4S6 | KIF21A | Kinesin-like protein KIF21A | Homo sapiens (Human) | EV |
| O75037 | KIF21B | Kinesin-like protein KIF21B | Homo sapiens (Human) | EV |
| Q9P2E2 | KIF17 | Kinesin-like protein KIF17 | Homo sapiens (Human) | EV |
| O00139 | KIF2A | Kinesin-like protein KIF2A | Homo sapiens (Human) | PR |
| Q9H1H9 | KIF13A | Kinesin-like protein KIF13A | Homo sapiens (Human) | SS |
| O43896 | KIF1C | Kinesin-like protein KIF1C | Homo sapiens (Human) | SS |
| Q9EQW7 | Kif13a | Kinesin-like protein KIF13A | Mus musculus (Mouse) | SS |
| Q70AM4 | Kif13b | Kinesin 13B | Rattus norvegicus (Rat) | EV |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGDSKVKVAV | RIRPMNRRET | DLHTKCVVDV | DANKVILNPV | NTNLSKGDAR | GQPKVFAYDH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CFWSMDESVK | EKYAGQDIVF | KCLGENILQN | AFDGYNACIF | AYGQTGSGKS | YTMMGTADQP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GLIPRLCSGL | FERTQKEENE | EQSFKVEVSY | MEIYNEKVRD | LLDPKGSRQT | LKVREHSVLG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PYVDGLSKLA | VTSYKDIESL | MSEGNKSRTV | AATNMNEESS | RSHAVFKITL | THTLYDVKSG |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TSGEKVGKLS | LVDLAGSERA | TKTGAAGDRL | KEGSNINKSL | TTLGLVISAL | ADQSAGKNKN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KFVPYRDSVL | TWLLKDSLGG | NSKTAMVATV | SPAADNYDET | LSTLRYADRA | KHIVNHAVVN |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EDPNARIIRD | LREEVEKLRE | QLTKAEAMKS | PELKDRLEES | EKLIQEMTVT | WEEKLRKTEE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| IAQERQKQLE | SLGISLQSSG | IKVGDDKCFL | VNLNADPALN | ELLVYYLKEH | TLIGSANSQD |
| 490 | 500 | 510 | 520 | 530 | 540 |
| IQLCGMGILP | EHCIIDITSE | GQVMLTPQKN | TRTFVNGSSV | SSPIQLHHGD | RILWGNNHFF |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RLNLPKKKKK | AEREDEDQDP | SMKNENSSEQ | LDVDGDSSSE | VSSEVNFNYE | YAQMEVTMKA |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LGSNDPMQSI | LNSLEQQHEE | EKRSALERQR | LMYEHELEQL | RRRLSPEKQN | CRSMDRFSFH |
| 670 | 680 | 690 | 700 | 710 | 720 |
| SPSAQQRLRQ | WAEEREATLN | NSLMRLREQI | VKANLLVREA | NYIAEELDKR | TEYKVTLQIP |
| 730 | 740 | 750 | 760 | 770 | 780 |
| ASSLDANRKR | GSLLSEPAIQ | VRRKGKGKQI | WSLEKLDNRL | LDMRDLYQEW | KECEEDNPVI |
| 790 | 800 | 810 | 820 | 830 | 840 |
| RSYFKRADPF | YDEQENHSLI | GVANVFLESL | FYDVKLQYAV | PIINQKGEVA | GRLHVEVMRL |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SGDVGERIAG | GDEVAEVSFE | KETQENKLVC | MVKILQATGL | PQHLSHFVFC | KYSFWDQQEP |
| 910 | 920 | 930 | 940 | 950 | 960 |
| VIVAPEVDTS | SSSVSKEPHC | MVVFDHCNEF | SVNITEDFIE | HLSEGALAIE | VYGHKINDPR |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| KNPALWDLGI | IQAKTRSLRD | RWSEVTRKLE | FWVQILEQNE | NGEYCPVEVI | SAKDVPTGGI |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| FQLRQGQSRR | VQVEVKSVQE | SGTLPLMEEC | ILSVGIGCVK | VRPLRAPRTH | ETFHEEEEDM |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| DSYQDRDLER | LRRKWLNALT | KRQEYLDQQL | QKLVSKRDKT | EDDADREAQL | LEMRLTLTEE |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| RNAVMVPSAG | SGIPGAPAEW | TPVPGMETHI | PVIFLDLNAD | DFSSQDNLDD | PEAGGWDATL |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| TGEEEEEFFE | LQIVKQHDGE | VKAEASWDSA | VHGCPQLSRG | TPVDERLFLI | VRVTVQLSHP |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| ADMQLVLRKR | ICVNVHGRQG | FAQSLLKKMS | HRSSIPGCGV | TFEIVSNIPE | DAQGVEEREA |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| LARMAANVEN | PASADSEAYI | EKYLRSVLAV | ENLLTLDRLR | QEVAVKEQLT | GKGKLSRRSI |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| SSPNVNRLSG | SRQDLIPSYS | LGSNKGRWES | QQDVSQTTVS | RGIAPAPALS | VSPQNNHSPD |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| PGLSNLAASY | LNPVKSFVPQ | MPKLLKSLFP | VRDEKRGKRP | SPLAHQPVPR | IMVQSASPDI |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| RVTRMEEAQP | EMGPDVLVQT | MGAPALKICD | KPAKVPSPPP | VIAVTAVTPA | PEAQDGPPSP |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| LSEASSGYFS | HSVSTATLSD | ALGPGLDAAA | PPGSMPTAPE | AEPEAPISHP | PPPTAVPAEE |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| PPGPQQLVSP | GRERPDLEAP | APGSPFRVRR | VRASELRSFS | RMLAGDPGCS | PGAEGNAPAP |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| GAGGQALASD | SEEADEVPEW | LREGEFVTVG | AHKTGVVRYV | GPADFQEGTW | VGVELDLPSG |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| KNDGSIGGKQ | YFRCNPGYGL | LVRPSRVRRA | TGPVRRRSTG | LRLGAPEARR | SATLSGSATN |
| 1810 | 1820 | ||||
| LASLTAALAK | ADRSHKNPEN | RKSWAS |