Q9MA88
Gene name |
MIRO3 |
Protein name |
Mitochondrial Rho GTPase 3 |
Names |
AtMIRO3, Miro-related GTPase 3 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G05310 |
EC number |
|
Protein Class |
|
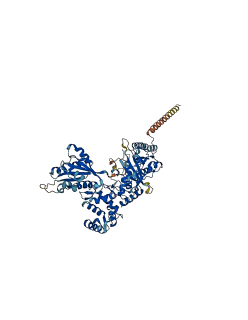
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9MA88
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9MA88-F1 | Predicted | AlphaFoldDB |
90 variants for Q9MA88
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_3_1510172_G_A | 5 | V>M | No | 1000Genomes | |
| tmp_3_1510185_G_A | 9 | S>N | No | 1000Genomes | |
| ENSVATH05774969 | 9 | S>R | No | 1000Genomes | |
| ENSVATH05774973 | 20 | V>I | No | 1000Genomes | |
| ENSVATH00307103 | 34 | A>T | No | 1000Genomes | |
| ENSVATH05774990 | 76 | G>E | No | 1000Genomes | |
| ENSVATH00307111 | 83 | R>K | No | 1000Genomes | |
| tmp_3_1510828_T_A | 92 | F>Y | No | 1000Genomes | |
| tmp_3_1510836_G_A | 95 | D>N | No | 1000Genomes | |
| ENSVATH05774991 | 110 | L>R | No | 1000Genomes | |
| ENSVATH05774995 | 144 | A>P | No | 1000Genomes | |
| ENSVATH02114584 | 144 | A>V | No | 1000Genomes | |
| ENSVATH05774997 | 145 | L>M | No | 1000Genomes | |
| ENSVATH05774998 | 147 | K>Q | No | 1000Genomes | |
| tmp_3_1511123_A_C | 149 | Y>S | No | 1000Genomes | |
| ENSVATH05775001 | 167 | K>N | No | 1000Genomes | |
| tmp_3_1511276_A_G | 167 | K>R | No | 1000Genomes | |
| ENSVATH05775002 | 168 | D>H | No | 1000Genomes | |
| ENSVATH05775002 | 168 | D>N | No | 1000Genomes | |
| ENSVATH05775003 | 177 | V>F | No | 1000Genomes | |
| ENSVATH02114586 | 196 | C>Y | No | 1000Genomes | |
| ENSVATH13872559 | 206 | L>R | No | 1000Genomes | |
| ENSVATH05775005 | 207 | S>Y | No | 1000Genomes | |
| tmp_3_1511401_C_T | 209 | H>Y | No | 1000Genomes | |
| ENSVATH05775007 | 216 | S>N | No | 1000Genomes | |
| ENSVATH05775010 | 226 | K>N | No | 1000Genomes | |
| ENSVATH00307113 | 242 | N>K | No | 1000Genomes | |
| ENSVATH02114589 | 246 | V>A | No | 1000Genomes | |
| ENSVATH00307114 | 247 | T>K | No | 1000Genomes | |
| tmp_3_1511598_A_T | 247 | T>S | No | 1000Genomes | |
| ENSVATH10502459 | 258 | T>K | No | 1000Genomes | |
| ENSVATH10502461 | 268 | R>H | No | 1000Genomes | |
| tmp_3_1511694_T_A | 279 | W>R | No | 1000Genomes | |
| ENSVATH00307115 | 288 | S>N | No | 1000Genomes | |
| ENSVATH05775012 | 294 | G>E | No | 1000Genomes | |
| ENSVATH02114590 | 305 | R>H | No | 1000Genomes | |
| ENSVATH13872561 | 307 | A>T | No | 1000Genomes | |
| ENSVATH02114591 | 307 | A>V | No | 1000Genomes | |
| ENSVATH10502464 | 315 | N>T | No | 1000Genomes | |
| tmp_3_1511942_G_A | 330 | S>N | No | 1000Genomes | |
| tmp_3_1511944_A_G | 331 | N>D | No | 1000Genomes | |
| ENSVATH05775017 | 337 | E>G | No | 1000Genomes | |
| ENSVATH05775018 | 341 | M>I | No | 1000Genomes | |
| ENSVATH00307120 | 357 | L>P | No | 1000Genomes | |
| ENSVATH05775022 | 357 | L>V | No | 1000Genomes | |
| ENSVATH10502531 | 362 | T>R | No | 1000Genomes | |
| ENSVATH10502532 | 363 | E>K | No | 1000Genomes | |
| ENSVATH00307121 | 366 | M>K | No | 1000Genomes | |
| ENSVATH02114602 | 379 | W>C | No | 1000Genomes | |
| ENSVATH00307122 | 388 | P>A | No | 1000Genomes | |
| ENSVATH00307123 | 408 | V>I | No | 1000Genomes | |
| ENSVATH13872563 | 410 | V>I | No | 1000Genomes | |
| ENSVATH13872563 | 410 | V>L | No | 1000Genomes | |
| tmp_3_1512459_A_T | 422 | K>N | No | 1000Genomes | |
| ENSVATH05775025 | 423 | S>Y | No | 1000Genomes | |
| tmp_3_1512464_A_G | 424 | E>G | No | 1000Genomes | |
| tmp_3_1512472_G_T | 427 | V>F | No | 1000Genomes | |
| ENSVATH10502581 | 435 | P>L | No | 1000Genomes | |
| ENSVATH05775026 | 449 | G>R | No | 1000Genomes | |
| ENSVATH05775027 | 454 | D>N | No | 1000Genomes | |
| tmp_3_1512583_G_A | 464 | D>N | No | 1000Genomes | |
| ENSVATH00307128 | 466 | H>R | No | 1000Genomes | |
| ENSVATH10502584 | 472 | V>G | No | 1000Genomes | |
| tmp_3_1512635_C_G | 481 | T>R | No | 1000Genomes | |
| ENSVATH00307129 | 490 | V>I | No | 1000Genomes | |
| ENSVATH10502615 | 506 | A>T | No | 1000Genomes | |
| ENSVATH05775028 | 508 | D>E | No | 1000Genomes | |
| ENSVATH05775029 | 515 | D>N | No | 1000Genomes | |
| ENSVATH05775030 | 516 | S>Y | No | 1000Genomes | |
| ENSVATH00307130 | 539 | S>A | No | 1000Genomes | |
| ENSVATH00307131 | 542 | V>L | No | 1000Genomes | |
| ENSVATH05775032 | 552 | T>I | No | 1000Genomes | |
| ENSVATH05775034 | 559 | V>M | No | 1000Genomes | |
| tmp_3_1512890_G_C | 566 | R>T | No | 1000Genomes | |
| tmp_3_1512982_C_G | 569 | Q>E | No | 1000Genomes | |
| ENSVATH10502616 | 571 | I>L | No | 1000Genomes | |
| tmp_3_1513019_C_A | 581 | S>Y | No | 1000Genomes | |
| tmp_3_1513023_A_C | 582 | K>N | No | 1000Genomes | |
| ENSVATH05775038 | 584 | G>R | No | 1000Genomes | |
| ENSVATH00307132 | 595 | T>I | No | 1000Genomes | |
| ENSVATH02114603 | 597 | A>T | No | 1000Genomes | |
| ENSVATH00307133 | 599 | N>T | No | 1000Genomes | |
| tmp_3_1513079_A_T | 601 | H>L | No | 1000Genomes | |
| ENSVATH05775041 | 602 | L>S | No | 1000Genomes | |
| ENSVATH00307134 | 615 | C>Y | No | 1000Genomes | |
| ENSVATH05775042 | 628 | G>R | No | 1000Genomes | |
| tmp_3_1513255_C_T | 634 | A>V | No | 1000Genomes | |
| ENSVATH05775043 | 637 | A>G | No | 1000Genomes | |
| tmp_3_1513273_G_A | 640 | R>H | No | 1000Genomes | |
| tmp_3_1513294_A_G | 647 | Q>R | No | 1000Genomes |
No associated diseases with Q9MA88
8 regional properties for Q9MA88
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | EF-hand domain | 316 - 351 | IPR002048 |
| domain | GTP binding domain | 430 - 522 | IPR006073 |
| domain | Mitochondrial Rho GTPase 1/3, EF hand associated, type-1 | 353 - 421 | IPR013566 |
| domain | EF hand associated, type-2 | 230 - 315 | IPR013567 |
| binding_site | EF-Hand 1, calcium-binding site | 208 - 220 | IPR018247-1 |
| binding_site | EF-Hand 1, calcium-binding site | 329 - 341 | IPR018247-2 |
| domain | MIRO domain | 12 - 179 | IPR020860-1 |
| domain | MIRO domain | 425 - 599 | IPR020860-2 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of mitochondrial outer membrane | The component of the mitochondrial outer membrane consisting of the gene products having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| mitochondrion organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a mitochondrion; includes mitochondrial morphogenesis and distribution, and replication of the mitochondrial genome as well as synthesis of new mitochondrial components. |
| regulation of mitochondrion organization | Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of a mitochondrion. |
| small GTPase mediated signal transduction | The series of molecular signals in which a small monomeric GTPase relays a signal. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P39722 | GEM1 | Mitochondrial Rho GTPase 1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q5ZM83 | RHOT2 | Mitochondrial Rho GTPase 2 | Gallus gallus (Chicken) | PR |
| Q8IMX7 | Miro | Mitochondrial Rho GTPase | Drosophila melanogaster (Fruit fly) | PR |
| Q9SU67 | ARAC8 | Rac-like GTP-binding protein ARAC8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82480 | ARAC7 | Rac-like GTP-binding protein ARAC7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38903 | ARAC2 | Rac-like GTP-binding protein ARAC2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82481 | ARAC10 | Rac-like GTP-binding protein ARAC10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MWMGVGDSSG | SPKPIRIVVV | GEKGSGKSSL | IMAAARNTFH | PNIPSLLPYT | NLPSEFFPDR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IPATVIDTSS | RPEDKGKVVK | EVRQADAIVL | TFAFDRPETL | DRLSKYWLPL | FRQLEVRVPI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IVAGYEVDNK | EAYNHFSIEQ | ITSALMKQYR | EVETSIQWSA | QRLDQAKDVL | YYAQKAVIDP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VGPVFDQENN | VLKPRCIAAL | KRIFLLSDHN | MDGILSDEEL | NELQKKCFDT | PLVPCEIKQM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KNVMQVTFPQ | GVNERGLTLD | GFLFLNTRLI | EEARIQTLWT | MLRKFGYSND | LRLGDDLVPY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SSFKRQADQS | VELTNVAIEF | LREVYEFFDS | NGDNNLEPHE | MGYLFETAPE | SPWTKPLYKD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VTEENMDGGL | SLEAFLSLWS | LMTLIDPPRS | LEYLMYIRFP | SDDPSSAVRV | TRKRVLDRKE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KKSERKVVQC | FVFGPKNAGK | SALLNQFIGR | SYDDDSNNNN | GSTDEHYAVN | MVKEPGVISD |
| 490 | 500 | 510 | 520 | 530 | 540 |
| TDKTLVLKEV | RIKDDGFMLS | KEALAACDVA | IFIYDSSDEY | SWNRAVDMLA | EVATIAKDSG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| YVFPCLMVAA | KTDLDPFPVA | IQESTRVTQD | IGIDAPIPIS | SKLGDVSNLF | RKILTAAENP |
| 610 | 620 | 630 | 640 | ||
| HLNIPEIESK | KKRSCKLNNR | SLMAVSIGTA | VLIAGLASFR | LYTARKQS |