Q9M2K0
Gene name |
XI-J (MYA3, MYOS3, XIJ, At3g58160, F9D24.70) |
Protein name |
Myosin-16 |
Names |
AtMYA3, AtMYOS3, Myosin XI J, AtXIJ |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G58160 |
EC number |
|
Protein Class |
|
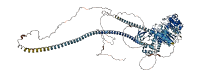
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9M2K0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9M2K0-F1 | Predicted | AlphaFoldDB |
70 variants for Q9M2K0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_3_21534807_A_T | 4 | N>I | No | 1000Genomes | |
| tmp_3_21534814_G_A | 6 | M>I | No | 1000Genomes | |
| ENSVATH12816513 | 11 | V>I | No | 1000Genomes | |
| ENSVATH12816514 | 29 | K>Q | No | 1000Genomes | |
| ENSVATH12816526 | 48 | S>P | No | 1000Genomes | |
| ENSVATH06336799 | 54 | D>Y | No | 1000Genomes | |
| ENSVATH00425669 | 67 | R>T | No | 1000Genomes | |
| tmp_3_21535331_G_A | 100 | V>I | No | 1000Genomes | |
| tmp_3_21535345_A_C | 104 | Q>H | No | 1000Genomes | |
| ENSVATH06336806 | 129 | H>Y | No | 1000Genomes | |
| ENSVATH12816530 | 205 | V>F | No | 1000Genomes | |
| tmp_3_21536203_C_A | 220 | Q>K | No | 1000Genomes | |
| tmp_3_21536209_G_C | 222 | D>H | No | 1000Genomes | |
| ENSVATH14450074 | 264 | E>V | No | 1000Genomes | |
| ENSVATH12816534 | 281 | N>K | No | 1000Genomes | |
| tmp_3_21536503_G_A | 296 | E>K | No | 1000Genomes | |
| ENSVATH14450105 | 306 | V>I | No | 1000Genomes | |
| tmp_3_21536542_A_T | 309 | I>L | No | 1000Genomes | |
| ENSVATH02520483 | 310 | S>N | No | 1000Genomes | |
| ENSVATH06336816 | 333 | S>L | No | 1000Genomes | |
| tmp_3_21536978_A_G | 402 | I>M | No | 1000Genomes | |
| tmp_3_21537121_C_T | 420 | Q>* | No | 1000Genomes | |
| tmp_3_21538344_C_G | 512 | P>R | No | 1000Genomes | |
| tmp_3_21538381_T_G | 524 | Y>* | No | 1000Genomes | |
| tmp_3_21538639_C_T | 581 | S>F | No | 1000Genomes | |
| ENSVATH12816587 | 584 | S>P | No | 1000Genomes | |
| tmp_3_21538660_C_T | 588 | P>L | No | 1000Genomes | |
| ENSVATH06336841 | 589 | P>A | No | 1000Genomes | |
| tmp_3_21538707_G_T | 604 | A>S | No | 1000Genomes | |
| ENSVATH06336844 | 619 | S>F | No | 1000Genomes | |
| tmp_3_21538955_C_A | 640 | D>E | No | 1000Genomes | |
| ENSVATH06336845 | 641 | N>K | No | 1000Genomes | |
| tmp_3_21539103_C_T | 665 | P>S | No | 1000Genomes | |
| tmp_3_21539148_C_T | 680 | L>F | No | 1000Genomes | |
| tmp_3_21539323_G_A | 703 | G>R | No | 1000Genomes | |
| tmp_3_21539329_A_T | 705 | T>S | No | 1000Genomes | |
| ENSVATH12816590 | 718 | A>V | No | 1000Genomes | |
| ENSVATH12816591 | 719 | G>C | No | 1000Genomes | |
| tmp_3_21539651_A_T | 761 | N>Y | No | 1000Genomes | |
| ENSVATH06336852 | 783 | A>S | No | 1000Genomes | |
| ENSVATH00425680 | 795 | I>V | No | 1000Genomes | |
| tmp_3_21539870_G_A | 801 | G>R | No | 1000Genomes | |
| ENSVATH06336854 | 803 | T>S | No | 1000Genomes | |
| ENSVATH02520498 | 824 | R>H | No | 1000Genomes | |
| ENSVATH12816594 | 828 | T>I | No | 1000Genomes | |
| ENSVATH06336862 | 877 | A>V | No | 1000Genomes | |
| tmp_3_21540322_A_G | 879 | R>G | No | 1000Genomes | |
| ENSVATH14450112 | 879 | R>K | No | 1000Genomes | |
| ENSVATH12816616 | 906 | H>Q | No | 1000Genomes | |
| tmp_3_21540403_C_T | 906 | H>Y | No | 1000Genomes | |
| tmp_3_21540416_G_T | 910 | S>I | No | 1000Genomes | |
| ENSVATH06336865 | 911 | D>G | No | 1000Genomes | |
| ENSVATH00425682 | 914 | E>Q | No | 1000Genomes | |
| tmp_3_21540469_G_A | 928 | E>K | No | 1000Genomes | |
| tmp_3_21540628_T_G | 981 | C>G | No | 1000Genomes | |
| ENSVATH12816617 | 1024 | F>I | No | 1000Genomes | |
| ENSVATH06336866 | 1043 | K>R | No | 1000Genomes | |
| ENSVATH02520500 | 1051 | G>V | No | 1000Genomes | |
| ENSVATH02520502 | 1052 | I>L | No | 1000Genomes | |
| tmp_3_21540901_C_G | 1072 | Q>E | No | 1000Genomes | |
| tmp_3_21540914_C_T | 1076 | P>L | No | 1000Genomes | |
| tmp_3_21541170_G_A | 1117 | G>R | No | 1000Genomes | |
| ENSVATH06336872 | 1128 | E>D | No | 1000Genomes | |
| tmp_3_21541430_C_G | 1160 | S>C | No | 1000Genomes | |
| tmp_3_21541438_G_A | 1163 | A>T | No | 1000Genomes | |
| tmp_3_21541439_C_T | 1163 | A>V | No | 1000Genomes | |
| ENSVATH06336877 | 1165 | T>I | No | 1000Genomes | |
| ENSVATH00425683 | 1184 | E>K | No | 1000Genomes | |
| ENSVATH12816622 | 1227 | T>I | No | 1000Genomes | |
| tmp_3_21541840_A_G | 1231 | D>G | No | 1000Genomes |
No associated diseases with Q9M2K0
5 regional properties for Q9M2K0
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| binding_site | IQ motif, EF-hand binding site | 731 - 753 | IPR000048-1 |
| binding_site | IQ motif, EF-hand binding site | 754 - 872 | IPR000048-2 |
| domain | Myosin head, motor domain | 54 - 730 | IPR001609 |
| domain | Myosin, N-terminal, SH3-like | 6 - 55 | IPR004009 |
| domain | Plant myosin, class XI, motor domain | 74 - 717 | IPR036018 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| myosin complex | A protein complex, formed of one or more myosin heavy chains plus associated light chains and other proteins, that functions as a molecular motor; uses the energy of ATP hydrolysis to move actin filaments or to move vesicles or other cargo on fixed actin filaments; has magnesium-ATPase activity and binds actin. Myosin classes are distinguished based on sequence features of the motor, or head, domain, but also have distinct tail regions that are believed to bind specific cargoes. |
| plant-type vacuole | A closed structure that is completely surrounded by a unit membrane, contains liquid, and retains the same shape regardless of cell cycle phase. An example of this structure is found in Arabidopsis thaliana. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| cytoskeletal motor activity | Generation of force resulting in movement, for example along a microfilament or microtubule, or in torque resulting in membrane scission or rotation of a flagellum. The energy required is obtained either from the hydrolysis of a nucleoside triphosphate or by an electrochemical proton gradient (proton-motive force). |
| hydrolase activity | Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. |
| microfilament motor activity | A motor activity that generates movement along a microfilament, driven by ATP hydrolysis. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| actin filament-based movement | Movement of organelles or other particles along actin filaments, or sliding of actin filaments past each other, mediated by motor proteins. |
| vesicle transport along actin filament | Movement of a vesicle along an actin filament, mediated by motor proteins. |
20 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q27991 | MYH10 | Myosin-10 | Bos taurus (Bovine) | SS |
| E1BPK6 | MYO6 | Unconventional myosin-VI | Bos taurus (Bovine) | SS |
| Q9I8D1 | MYO6 | Unconventional myosin-VI | Gallus gallus (Chicken) | SS |
| Q02440 | MYO5A | Unconventional myosin-Va | Gallus gallus (Chicken) | SS |
| Q5U651 | RASIP1 | Ras-interacting protein 1 | Homo sapiens (Human) | PR |
| B0I1T2 | MYO1G | Unconventional myosin-Ig | Homo sapiens (Human) | PR |
| P35580 | MYH10 | Myosin-10 | Homo sapiens (Human) | SS |
| Q9UM54 | MYO6 | Unconventional myosin-VI | Homo sapiens (Human) | EV |
| Q9Y4I1 | MYO5A | Unconventional myosin-Va | Homo sapiens (Human) | SS |
| Q9NQX4 | MYO5C | Unconventional myosin-Vc | Homo sapiens (Human) | SS |
| Q9ULV0 | MYO5B | Unconventional myosin-Vb | Homo sapiens (Human) | SS |
| Q3U0S6 | Rasip1 | Ras-interacting protein 1 | Mus musculus (Mouse) | PR |
| Q61879 | Myh10 | Myosin-10 | Mus musculus (Mouse) | SS |
| Q99104 | Myo5a | Unconventional myosin-Va | Mus musculus (Mouse) | EV |
| P21271 | Myo5b | Unconventional myosin-Vb | Mus musculus (Mouse) | SS |
| Q64331 | Myo6 | Unconventional myosin-VI | Mus musculus (Mouse) | SS |
| Q29122 | MYO6 | Unconventional myosin-VI | Sus scrofa (Pig) | SS |
| Q9JLT0 | Myh10 | Myosin-10 | Rattus norvegicus (Rat) | SS |
| Q9QYF3 | Myo5a | Unconventional myosin-Va | Rattus norvegicus (Rat) | SS |
| P70569 | Myo5b | Unconventional myosin-Vb | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAENIMVDSH | VWVEDPERAW | IDGVVLNIKG | EEAEIKTNDG | RDVIANLSRL | YPKDTEAPSE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GVEDMTRLSY | LHEPAVLDNL | ATRYELNEIY | TYTGNILIAV | NPFQGLPHLY | DAEVMEKYKE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AYFKELNPHV | FAIGGIAYRE | MINEGRNKCI | LVSGESGSGK | TETTKMLMRY | LAYFGGHTAV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EGRTVENQVL | ESNPVLEAFG | NAKTVKNNNS | SRFGKFVEIQ | FDDVGRISGA | AIRTYLLERS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RVCQVSDPER | NYHCFYLLCA | APPEDVERFK | LGDPKSFRYL | NQSSCYKLDG | VNDAEEYLAT |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RRAMDVVGIS | EKEQDAIFRV | VASILHLGNI | EFSKGEDADS | SSVKDEQSMF | HLQMTSELLM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| CDPHSLEDAL | CKRMMVTPEE | VIKRSLDPLG | AAVSRDGLAK | TIYSRLFDWL | VNKINISIGQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| DSHSRRLIGV | LDIYGFESFK | TNSFEQFCIN | YTNEKLQQHF | NQHVFKMEQG | EYQKEEIDWS |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YVEFVDNKDV | VDLIEKKPGG | IIALLDEACM | LPKSTPETFS | EKLYHTFKDH | KRFMKPKLTR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SDFTLVHYAG | DVQYQSDQFL | DKNKDYVVAE | HQDLLNASKC | SFVSGLFPPL | PKESSKSKFS |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SIGARFKLQL | QQLMETLNST | EPHYIRCVKP | NNLLQPTVFD | NANVLHQLRS | GGVLEAIRVK |
| 670 | 680 | 690 | 700 | 710 | 720 |
| CAGYPTNRTF | IEFLNRFLIL | APEILKGEYE | AEVACKWILE | KKGLTGYQIG | KSKVFLRAGQ |
| 730 | 740 | 750 | 760 | 770 | 780 |
| MAELDAHRTR | VLGESARMIQ | GQVRTRLTRE | RFVLMRRASV | NIQANWRGNI | ARKISKEMRR |
| 790 | 800 | 810 | 820 | 830 | 840 |
| EEAAIKIQKN | LRRQIAKKDY | GKTKSSALTL | QSGVRTMAAR | HEFRYKLTTR | AATVIQAYWR |
| 850 | 860 | 870 | 880 | 890 | 900 |
| GYSAISDYKK | LKRVSLLCKS | NLRGRIARKQ | LGQSKQADRK | EETEKERKVE | LSNRAEEAVD |
| 910 | 920 | 930 | 940 | 950 | 960 |
| MSFVLHSEQS | DDAESGHGRK | AKLSIESEDG | LDKSSVLHSE | QSDDEELGHE | RKTKLSIESE |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| DGHSDQSDDE | EIEHERKTKH | CIQAEDGIEK | SYVMHSDQSD | DEEIGHKRKT | KHSIQAEDGI |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| EKSFVVHSDQ | SDDEEIGHER | KTKHAIQVED | GIQKSFVTCS | EKPYNTFSVV | SQITSPIRDT |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| EIESLTAEVE | MLKALLQVEK | QRADISERKC | AEARELGERR | RKRLEETERR | VYQLQDSLNR |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| LLYSMSDQFS | QLKSILRSPS | MSASTMASAP | VVRDDLADSS | ENSEASSSDS | DFTFPAPSPS |
| 1210 | 1220 | 1230 | 1240 | ||
| SDNFSTFNPN | QLQVIVQDLS | TTEAKGTESY | DSDKEGGFED | YF |