Q9M0Y8
Gene name |
NSF |
Protein name |
Vesicle-fusing ATPase |
Names |
N-ethylmaleimide-sensitive fusion protein, Vesicular-fusion protein NSF |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT4G04910 |
EC number |
3.6.4.6: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
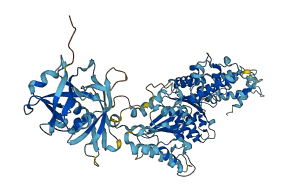
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
249-396 (D1 domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9M0Y8
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9M0Y8-F1 | Predicted | AlphaFoldDB |
14 variants for Q9M0Y8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_4_2495659_C_T | 3 | G>D | No | 1000Genomes | |
| tmp_4_2495656_C_T | 4 | R>H | No | 1000Genomes | |
| ENSVATH13942758 | 97 | N>I | No | 1000Genomes | |
| ENSVATH06477324 | 130 | R>T | No | 1000Genomes | |
| ENSVATH00464834 | 159 | A>V | No | 1000Genomes | |
| ENSVATH10753989 | 165 | D>H | No | 1000Genomes | |
| ENSVATH10753988 | 185 | A>S | No | 1000Genomes | |
| ENSVATH06477275 | 462 | P>S | No | 1000Genomes | |
| ENSVATH06477268 | 547 | A>G | No | 1000Genomes | |
| tmp_4_2489993_T_C | 677 | N>S | No | 1000Genomes | |
| ENSVATH10753772 | 722 | R>K | No | 1000Genomes | |
| ENSVATH02661867 | 723 | E>G | No | 1000Genomes | |
| ENSVATH02661866 | 724 | K>R | No | 1000Genomes | |
| tmp_4_2489697_T_A | 743 | G>L | No | 1000Genomes |
No associated diseases with Q9M0Y8
8 regional properties for Q9M0Y8
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | CDC48, N-terminal subdomain | 11 - 95 | IPR003338 |
| domain | AAA+ ATPase domain | 249 - 396 | IPR003593-1 |
| domain | AAA+ ATPase domain | 529 - 650 | IPR003593-2 |
| domain | ATPase, AAA-type, core | 253 - 393 | IPR003959-1 |
| domain | ATPase, AAA-type, core | 534 - 644 | IPR003959-2 |
| conserved_site | ATPase, AAA-type, conserved site | 364 - 382 | IPR003960 |
| domain | CDC48, domain 2 | 118 - 166 | IPR004201 |
| domain | AAA ATPase, AAA+ lid domain | 420 - 475 | IPR041569 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.6 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| Golgi stack | The set of thin, flattened membrane-bounded compartments, called cisternae, that form the central portion of the Golgi complex. The stack usually comprises cis, medial, and trans cisternae; the cis- and trans-Golgi networks are not considered part of the stack. |
| plant-type vacuole | A closed structure that is completely surrounded by a unit membrane, contains liquid, and retains the same shape regardless of cell cycle phase. An example of this structure is found in Arabidopsis thaliana. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| plasmodesma | A fine cytoplasmic channel, found in all higher plants, that connects the cytoplasm of one cell to that of an adjacent cell. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| metal ion binding | Binding to a metal ion. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| Golgi organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the Golgi apparatus. |
| Golgi to plasma membrane protein transport | The directed movement of proteins from the Golgi to the plasma membrane in transport vesicles that move from the trans-Golgi network to the plasma membrane. |
| Golgi vesicle docking | The initial attachment of a Golgi transport vesicle membrane to a target membrane, mediated by proteins protruding from the membrane of the Golgi vesicle and the target membrane. |
| intra-Golgi vesicle-mediated transport | The directed movement of substances within the Golgi, mediated by small transport vesicles. These either fuse with the cis-Golgi or with each other to form the membrane stacks known as the cis-Golgi reticulum (network). |
| SNARE complex disassembly | The disaggregation of the SNARE protein complex into its constituent components. The SNARE complex is a protein complex involved in membrane fusion; a stable ternary complex consisting of a four-helix bundle, usually formed from one R-SNARE and three Q-SNAREs with an ionic layer sandwiched between hydrophobic layers. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P46461 | comt | Vesicle-fusing ATPase 1 | Drosophila melanogaster (Fruit fly) | SS |
| P54351 | Nsf2 | Vesicle-fusing ATPase 2 | Drosophila melanogaster (Fruit fly) | SS |
| P46459 | NSF | Vesicle-fusing ATPase | Homo sapiens (Human) | SS |
| P46460 | Nsf | Vesicle-fusing ATPase | Mus musculus (Mouse) | SS |
| Q9QUL6 | Nsf | Vesicle-fusing ATPase | Rattus norvegicus (Rat) | SS |
| Q94392 | nsf-1 | Vesicle-fusing ATPase | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAGRYGSQVM | TMTVTNTPSA | DLAFTNLAYC | SSSDLRQFSV | PGSDLFLANV | ADSFILSLCG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| HGSIRDGNIA | LNAIQRRHAR | VSTGDMVSVS | RFVPPENFDL | AMLTLELEFV | KKGTKSEQVD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AALLSTQLKR | KYTNQVLTVG | QKATFEYHGT | NYILTVNRAD | VEGQDHTNGI | ERGLLSKDTY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IVFEASNASG | IKIVNQREAA | SSNIFKHKEF | NLESLGIGGL | GAEFADIFRR | AFASRVFPPH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VTSRLGIKHV | KGMLLFGPPG | TGKTLMARQI | GKMLNGKDPK | IVNGPEVLSK | FVGETEKNVR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DLFADAEQDQ | RTLGDASELH | VIIFDEIDAI | CKSRGSTRDG | TGVHDSIVNQ | LLTKIDGVEA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LNNVLLIGMT | NRKDLLDEAL | LRPGRLEVQV | EISLPDEAGR | LQILQIHTNK | MKENSFLGTD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| INLQELAART | KNYSGAELEG | VVKSATSYAL | NRQLSMDDLT | KPVEEENIKI | TMEDFLHAIY |
| 490 | 500 | 510 | 520 | 530 | 540 |
| EVQPAFGAST | DDLERCRLNG | MVDCGHRHNH | IYKRAMLLVE | QVKVSTRSPL | VTCLLEGPSG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SGKTALAATI | GIDSDFPYVK | IVSAETMIGL | SESTKCAHIV | KVFEDAYKSP | MSIIILDDIE |
| 610 | 620 | 630 | 640 | 650 | 660 |
| RLLEFIAIGP | RFSNIISQTL | MVLLKRLPPK | GKKLLVFGTT | SEVTFLESVG | ISDCFSVTHS |
| 670 | 680 | 690 | 700 | 710 | 720 |
| VPTLQKEDAK | KVLNQLNLFS | EDDVDSAAEA | LNDMPIKKIY | MLIEMAAQGE | NGGSAEAIYA |
| 730 | 740 | ||||
| GREKININHF | YDCLGDFIRF | TG |