Q9LYN6
Gene name |
At3g56050 (F18O21.10) |
Protein name |
Probable inactive receptor-like protein kinase At3g56050 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G56050 |
EC number |
|
Protein Class |
|
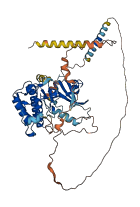
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9LYN6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9LYN6-F1 | Predicted | AlphaFoldDB |
36 variants for Q9LYN6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH06324609 | 2 | S>T | No | 1000Genomes | |
| ENSVATH06324608 | 17 | L>F | No | 1000Genomes | |
| ENSVATH14441934 | 30 | Y>F | No | 1000Genomes | |
| ENSVATH02512739 | 30 | Y>H | No | 1000Genomes | |
| tmp_3_20800508_T_A | 37 | D>V | No | 1000Genomes | |
| ENSVATH06324605 | 52 | D>A | No | 1000Genomes | |
| tmp_3_20800455_C_T | 55 | D>N | No | 1000Genomes | |
| ENSVATH06324595 | 81 | D>G | No | 1000Genomes | |
| tmp_3_20799874_G_T | 83 | P>T | No | 1000Genomes | |
| ENSVATH06324594 | 94 | V>I | No | 1000Genomes | |
| tmp_3_20799834_G_A | 96 | T>M | No | 1000Genomes | |
| tmp_3_20799831_C_T | 97 | R>K | No | 1000Genomes | |
| ENSVATH02512723 | 99 | D>N | No | 1000Genomes | |
| tmp_3_20799774_T_A | 116 | Q>L | No | 1000Genomes | |
| tmp_3_20799768_A_C | 118 | V>G | No | 1000Genomes | |
| tmp_3_20799681_A_G | 147 | I>T | No | 1000Genomes | |
| tmp_3_20799682_T_C | 147 | I>V | No | 1000Genomes | |
| ENSVATH06324590 | 148 | V>L | No | 1000Genomes | |
| ENSVATH06324590 | 148 | V>M | No | 1000Genomes | |
| ENSVATH00422064 | 152 | I>V | No | 1000Genomes | |
| ENSVATH12787782 | 161 | L>S | No | 1000Genomes | |
| tmp_3_20799614_T_G | 169 | K>N | No | 1000Genomes | |
| ENSVATH14441931 | 172 | A>V | No | 1000Genomes | |
| tmp_3_20799252_A_C | 248 | N>K | No | 1000Genomes | |
| ENSVATH12787779 | 250 | I>L | No | 1000Genomes | |
| tmp_3_20799137_T_A | 259 | E>D | No | 1000Genomes | |
| tmp_3_20799028_C_T | 296 | V>I | No | 1000Genomes | |
| tmp_3_20798886_C_G | 312 | M>I | No | 1000Genomes | |
| ENSVATH00422059 | 313 | R>Q | No | 1000Genomes | |
| tmp_3_20798819_C_G | 335 | V>L | No | 1000Genomes | |
| tmp_3_20798767_A_G | 352 | V>A | No | 1000Genomes | |
| ENSVATH02512700 | 400 | M>I | No | 1000Genomes | |
| tmp_3_20798602_G_A | 407 | S>L | No | 1000Genomes | |
| ENSVATH02512697 | 409 | Q>K | No | 1000Genomes | |
| ENSVATH12787764 | 412 | D>E | No | 1000Genomes | |
| tmp_3_20798464_A_G | 453 | I>T | No | 1000Genomes |
No associated diseases with Q9LYN6
4 regional properties for Q9LYN6
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
25 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9Y616 | IRAK3 | Interleukin-1 receptor-associated kinase 3 | Homo sapiens (Human) | PR |
| P53667 | LIMK1 | LIM domain kinase 1 | Homo sapiens (Human) | PR |
| P53671 | LIMK2 | LIM domain kinase 2 | Homo sapiens (Human) | PR |
| Q8K4B2 | Irak3 | Interleukin-1 receptor-associated kinase 3 | Mus musculus (Mouse) | PR |
| Q8CFA1 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Mus musculus (Mouse) | PR |
| O54785 | Limk2 | LIM domain kinase 2 | Mus musculus (Mouse) | PR |
| Q4QQS0 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Rattus norvegicus (Rat) | PR |
| Q9ASQ5 | CRCK3 | Calmodulin-binding receptor-like cytoplasmic kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STF0 | LECRKS3 | Receptor like protein kinase S.3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCZ4 | FER | Receptor-like protein kinase FERONIA | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VZJ9 | CRCK2 | Calmodulin-binding receptor-like cytoplasmic kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94C25 | At5g20050 | Probable receptor-like protein kinase At5g20050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FLW0 | At5g24010 | Probable receptor-like protein kinase At5g24010 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LTC0 | PBL19 | Probable serine/threonine-protein kinase PBL19 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FIL7 | CRCK1 | Calmodulin-binding receptor-like cytoplasmic kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGQ4 | MDIS2 | Protein MALE DISCOVERER 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSNNWKSVRL | RLQNRTLVFL | LVILSFGSCY | SLKSQGDGFL | ESVTKDLWSD | IDAEDLRAVG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FHRKLLGRFR | NPYTHLNAFR | DRPVARATPP | SSSVSTRPDA | KRSSTLPPPQ | KSPPAQHVSA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PPPFVHHVTL | PSLTSSSKTS | SNSTIPIVAG | CIAGAVFILL | LATGVFFFKS | KAGKSVNPWR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TGLSGQLQKV | FITGVPKLKR | SEIEAACEDF | SNVIGSCPIG | TLFKGTLSSG | VEIAVASVAT |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ASAKEWTNNI | EMQFRKKIEM | LSKINHKNFV | NLLGYCEEEE | PFTRILVFEY | ASNGTVFEHL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HYKESEHLDW | VMRLRIAMGI | AYCLDHMHGL | KPPIVHSNLL | SSSVQLTEDY | AVKIADFNFG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| YLKGPSETES | STNALIDTNI | SETTQEDNVH | SFGLLLFELM | TGKLPESVQK | GDSIDTGLAV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| FLRGKTLREM | VDPTIESFDE | KIENIGEVIK | SCIRADAKQR | PIMKEVTGRL | REITGLSPDD |
| 490 | |||||
| TIPKLSPLWW | AELEVLSTA |