Q9LT35
Gene name |
NEK6 (At3g20860, MOE17.17) |
Protein name |
Serine/threonine-protein kinase Nek6 |
Names |
NimA-related protein kinase 6, AtNek6 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G20860 |
EC number |
2.7.11.34: Protein-serine/threonine kinases |
Protein Class |
|
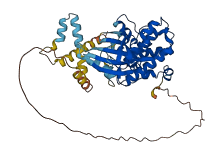
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
145-167 (Activation loop from InterPro)
Target domain |
4-257 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9LT35
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9LT35-F1 | Predicted | AlphaFoldDB |
25 variants for Q9LT35
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH05872618 | 7 | V>L | No | 1000Genomes | |
| tmp_3_7306232_C_T | 18 | A>V | No | 1000Genomes | |
| tmp_3_7306264_A_T | 29 | K>* | No | 1000Genomes | |
| ENSVATH05872621 | 31 | V>L | No | 1000Genomes | |
| ENSVATH05872626 | 81 | S>G | No | 1000Genomes | |
| ENSVATH02170995 | 96 | R>K | No | 1000Genomes | |
| ENSVATH00336924 | 98 | V>I | No | 1000Genomes | |
| tmp_3_7307408_T_C | 188 | L>S | No | 1000Genomes | |
| tmp_3_7307489_T_A | 190 | C>S | No | 1000Genomes | |
| tmp_3_7307672_A_C | 225 | M>L | No | 1000Genomes | |
| ENSVATH05872650 | 278 | S>L | No | 1000Genomes | |
| ENSVATH02171006 | 282 | T>N | No | 1000Genomes | |
| ENSVATH02171007 | 289 | L>I | No | 1000Genomes | |
| tmp_3_7308078_C_A | 299 | P>T | No | 1000Genomes | |
| ENSVATH02171011 | 306 | S>* | No | 1000Genomes | |
| ENSVATH02171010 | 306 | S>A | No | 1000Genomes | |
| ENSVATH02171013 | 312 | K>I | No | 1000Genomes | |
| ENSVATH02171013 | 312 | K>R | No | 1000Genomes | |
| ENSVATH05872651 | 316 | T>A | No | 1000Genomes | |
| ENSVATH10813590 | 325 | R>I | No | 1000Genomes | |
| ENSVATH02171014 | 327 | A>T | No | 1000Genomes | |
| tmp_3_7308235_G_T | 351 | S>I | No | 1000Genomes | |
| tmp_3_7308283_G_T | 367 | G>V | No | 1000Genomes | |
| ENSVATH05872654 | 405 | V>I | No | 1000Genomes | |
| ENSVATH02171018 | 416 | I>L | No | 1000Genomes |
No associated diseases with Q9LT35
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.34 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cortical microtubule organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of structures formed of microtubules and associated proteins in the cell cortex, i.e. just beneath the plasma membrane of a cell. |
| lateral root formation | The process that gives rise to a lateral root. This process pertains to the initial formation of a structure from unspecified parts. A lateral root primordium represents an organized group of cells derived from the root pericycle that will differentiate into a new root, as opposed to the initiation of the main root from the embryo proper. |
| negative regulation of ethylene biosynthetic process | Any process that stops, prevents, or reduces the frequency, rate or extent of an ethylene biosynthetic process. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P22209 | KIN3 | Serine/threonine-protein kinase KIN3 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q6P3R8 | NEK5 | Serine/threonine-protein kinase Nek5 | Homo sapiens (Human) | PR |
| Q8NE28 | STKLD1 | Serine/threonine kinase-like domain-containing protein STKLD1 | Homo sapiens (Human) | PR |
| P51957 | NEK4 | Serine/threonine-protein kinase Nek4 | Homo sapiens (Human) | PR |
| P51955 | NEK2 | Serine/threonine-protein kinase Nek2 | Homo sapiens (Human) | EV |
| Q8K1R7 | Nek9 | Serine/threonine-protein kinase Nek9 | Mus musculus (Mouse) | SS |
| Q7TSC3 | Nek5 | Serine/threonine-protein kinase Nek5 | Mus musculus (Mouse) | PR |
| O35942 | Nek2 | Serine/threonine-protein kinase Nek2 | Mus musculus (Mouse) | PR |
| Q10GB1 | NEK1 | Serine/threonine-protein kinase Nek1 | Oryza sativa subsp japonica (Rice) | PR |
| Q6ZEZ5 | NEK3 | Serine/threonine-protein kinase Nek3 | Oryza sativa subsp japonica (Rice) | PR |
| Q8RXT4 | NEK4 | Serine/threonine-protein kinase Nek4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SLI2 | NEK1 | Serine/threonine-protein kinase Nek1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDDYEVVEQI | GRGAFGSAFL | VIHKSERRKY | VVKKIRLAKQ | TERCKLAAIQ | EMSLISKLKS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PYIVEYKDSW | VEKDCVCIVT | SYCEGGDMTQ | MIKKSRGVFA | SEEKLCRWMV | QLLLAIDYLH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NNRVLHRDLK | CSNIFLTKEN | EVRLGDFGLA | KLLGKDDLAS | SMVGTPNYMC | PELLADIPYG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YKSDIWSLGC | CMFEVAAHQP | AFKAPDMAAL | INKINRSSLS | PLPVMYSSSL | KRLIKSMLRK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NPEHRPTAAE | LLRHPHLQPY | LAQCQNLSPV | FKPVVSKSEH | NTNENRTGLP | PKTKSAKTPI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KHNQESEETE | KKNKDTSSSS | KDKERPAKSQ | EMSVISTLTL | LREFQKKSPK | SEERAEALES |
| 370 | 380 | 390 | 400 | 410 | |
| LLELCAGLLR | QEKFDELEGV | LKPFGDETVS | SRETAIWLTK | SLMNVKRKQN | DDETNI |