Q9LSV3
Gene name |
WAKL16 (At3g25490, MWL2.11) |
Protein name |
Putative wall-associated receptor kinase-like 16 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G25490 |
EC number |
|
Protein Class |
|
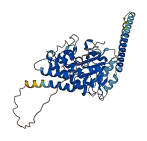
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
250-274 (Activation loop from InterPro)
Target domain |
108-391 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9LSV3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9LSV3-F1 | Predicted | AlphaFoldDB |
122 variants for Q9LSV3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH10975107 | 3 | L>V | No | 1000Genomes | |
| ENSVATH02201753 | 6 | V>I | No | 1000Genomes | |
| tmp_3_9241744_T_G | 7 | V>G | No | 1000Genomes | |
| ENSVATH02201754 | 8 | Y>* | No | 1000Genomes | |
| ENSVATH02201756 | 10 | V>L | No | 1000Genomes | |
| ENSVATH10975108 | 11 | A>S | No | 1000Genomes | |
| tmp_3_9241766_C_A | 14 | F>L | No | 1000Genomes | |
| ENSVATH00349292 | 21 | I>F | No | 1000Genomes | |
| ENSVATH02201758 | 22 | A>G | No | 1000Genomes | |
| tmp_3_9241794_A_C,G | 24 | I>L | No | 1000Genomes | |
| ENSVATH05912702 | 24 | I>M | No | 1000Genomes | |
| tmp_3_9241794_A_C,G | 24 | I>V | No | 1000Genomes | |
| tmp_3_9241807_A_T | 28 | K>M | No | 1000Genomes | |
| ENSVATH02201759 | 28 | K>N | No | 1000Genomes | |
| tmp_3_9241814_A_T | 30 | L>F | No | 1000Genomes | |
| ENSVATH00349293 | 34 | M>I | No | 1000Genomes | |
| ENSVATH05912703 | 35 | I>V | No | 1000Genomes | |
| ENSVATH14005346 | 36 | I>V | No | 1000Genomes | |
| ENSVATH05912709 | 45 | S>N | No | 1000Genomes | |
| tmp_3_9241981_G_C | 57 | R>T | No | 1000Genomes | |
| ENSVATH05912710 | 59 | H>Y | No | 1000Genomes | |
| ENSVATH05912711 | 62 | D>N | No | 1000Genomes | |
| ENSVATH02201763 | 70 | F>V | No | 1000Genomes | |
| tmp_3_9242027_GA_TA,G | 72 | E>D | No | 1000Genomes | |
| ENSVATH05912712 | 75 | G>S | No | 1000Genomes | |
| tmp_3_9242046_T_A | 79 | L>M | No | 1000Genomes | |
| ENSVATH00349297 | 83 | L>V | No | 1000Genomes | |
| ENSVATH02201768 | 89 | S>P | No | 1000Genomes | |
| ENSVATH00349298 | 96 | F>L | No | 1000Genomes | |
| ENSVATH05912713 | 97 | T>M | No | 1000Genomes | |
| ENSVATH10975113 | 105 | T>A | No | 1000Genomes | |
| ENSVATH02201770 | 107 | G>D | No | 1000Genomes | |
| ENSVATH10975114 | 107 | G>S | No | 1000Genomes | |
| ENSVATH00349299 | 112 | R>K | No | 1000Genomes | |
| ENSVATH00349300 | 117 | G>R | No | 1000Genomes | |
| ENSVATH10975156 | 118 | G>S | No | 1000Genomes | |
| tmp_3_9242167_A_T | 119 | Q>L | No | 1000Genomes | |
| ENSVATH00349301 | 120 | W>G | No | 1000Genomes | |
| tmp_3_9242176_T_G | 122 | V>G | No | 1000Genomes | |
| ENSVATH10975157 | 128 | P>L | No | 1000Genomes | |
| ENSVATH02201772 | 129 | D>H | No | 1000Genomes | |
| ENSVATH02201772 | 129 | D>N | No | 1000Genomes | |
| ENSVATH00349302 | 130 | N>K | No | 1000Genomes | |
| tmp_3_9242209_T_C | 133 | V>A | No | 1000Genomes | |
| ENSVATH05912715 | 133 | V>F | No | 1000Genomes | |
| tmp_3_9242248_T_A | 146 | V>E | No | 1000Genomes | |
| ENSVATH05912717 | 151 | N>D | No | 1000Genomes | |
| ENSVATH02201773 | 164 | V>F | No | 1000Genomes | |
| ENSVATH02201774 | 167 | L>V | No | 1000Genomes | |
| tmp_3_9242354_T_G | 181 | Y>* | No | 1000Genomes | |
| ENSVATH00349303 | 185 | T>S | No | 1000Genomes | |
| ENSVATH02201777 | 191 | D>N | No | 1000Genomes | |
| ENSVATH02201777 | 191 | D>Y | No | 1000Genomes | |
| tmp_3_9242394_G_C | 195 | G>R | No | 1000Genomes | |
| ENSVATH00349304 | 197 | M>L | No | 1000Genomes | |
| ENSVATH00349305 | 199 | V>A | No | 1000Genomes | |
| ENSVATH05912719 | 207 | R>L | No | 1000Genomes | |
| tmp_3_9242442_G_A | 211 | A>T | No | 1000Genomes | |
| ENSVATH05912720 | 216 | G>R | No | 1000Genomes | |
| ENSVATH00349307 | 218 | I>L | No | 1000Genomes | |
| ENSVATH00349307 | 218 | I>V | No | 1000Genomes | |
| ENSVATH10975160 | 222 | H>P | No | 1000Genomes | |
| ENSVATH00349309 | 222 | H>Y | No | 1000Genomes | |
| ENSVATH02201779 | 232 | R>C | No | 1000Genomes | |
| ENSVATH02201780 | 232 | R>H | No | 1000Genomes | |
| ENSVATH02201780 | 232 | R>L | No | 1000Genomes | |
| ENSVATH02201779 | 232 | R>S | No | 1000Genomes | |
| ENSVATH02201782 | 243 | E>K | No | 1000Genomes | |
| ENSVATH05912721 | 244 | N>T | No | 1000Genomes | |
| ENSVATH05912722 | 245 | L>I | No | 1000Genomes | |
| ENSVATH10975161 | 254 | A>V | No | 1000Genomes | |
| ENSVATH00349310 | 258 | K>M | No | 1000Genomes | |
| ENSVATH10975164 | 259 | P>S | No | 1000Genomes | |
| ENSVATH05912724 | 268 | M>I | No | 1000Genomes | |
| ENSVATH00349312 | 276 | L>V | No | 1000Genomes | |
| ENSVATH00349313 | 285 | L>I | No | 1000Genomes | |
| ENSVATH02201785 | 289 | K>N | No | 1000Genomes | |
| ENSVATH05912726 | 291 | D>V | No | 1000Genomes | |
| tmp_3_9242711_G_T | 300 | M>I | No | 1000Genomes | |
| tmp_3_9242712_G_A | 301 | E>K | No | 1000Genomes | |
| tmp_3_9242724_G_C | 305 | G>R | No | 1000Genomes | |
| ENSVATH02201787 | 307 | K>Q | No | 1000Genomes | |
| ENSVATH10975196 | 312 | E>A | No | 1000Genomes | |
| ENSVATH02201788 | 315 | E>D | No | 1000Genomes | |
| tmp_3_9242766_C_A | 319 | H>N | No | 1000Genomes | |
| ENSVATH05912728 | 320 | L>S | No | 1000Genomes | |
| tmp_3_9242772_G_A | 321 | V>M | No | 1000Genomes | |
| ENSVATH00349314 | 325 | V>L | No | 1000Genomes | |
| ENSVATH10975197 | 326 | L>F | No | 1000Genomes | |
| ENSVATH10975198 | 335 | E>V | No | 1000Genomes | |
| ENSVATH00349315 | 345 | E>D | No | 1000Genomes | |
| tmp_3_9242849_C_A | 346 | N>K | No | 1000Genomes | |
| tmp_3_9242860_T_G | 350 | I>S | No | 1000Genomes | |
| tmp_3_9242872_C_T | 354 | A>V | No | 1000Genomes | |
| ENSVATH05912729 | 356 | V>D | No | 1000Genomes | |
| ENSVATH00349317 | 358 | V>L | No | 1000Genomes | |
| ENSVATH00349318 | 359 | E>D | No | 1000Genomes | |
| tmp_3_9242896_G_A | 362 | R>K | No | 1000Genomes | |
| ENSVATH02201792 | 363 | L>P | No | 1000Genomes | |
| ENSVATH02201795 | 367 | E>D | No | 1000Genomes | |
| tmp_3_9242910_G_A | 367 | E>K | No | 1000Genomes | |
| ENSVATH05912730 | 368 | R>G | No | 1000Genomes | |
| ENSVATH14005347 | 369 | P>S | No | 1000Genomes | |
| tmp_3_9242929_A_C | 373 | E>A | No | 1000Genomes | |
| ENSVATH00349319 | 375 | A>V | No | 1000Genomes | |
| tmp_3_9242940_G_A | 377 | E>K | No | 1000Genomes | |
| tmp_3_9242986_A_T | 392 | D>V | No | 1000Genomes | |
| ENSVATH02201797 | 395 | P>L | No | 1000Genomes | |
| ENSVATH02201796 | 395 | P>T | No | 1000Genomes | |
| ENSVATH00349324 | 399 | V>E | No | 1000Genomes | |
| ENSVATH02201799 | 401 | L>F | No | 1000Genomes | |
| tmp_3_9243022_G_T | 404 | S>I | No | 1000Genomes | |
| ENSVATH00349326 | 407 | V>L | No | 1000Genomes | |
| ENSVATH00349327 | 410 | Q>P | No | 1000Genomes | |
| ENSVATH00349328 | 411 | G>S | No | 1000Genomes | |
| ENSVATH00349329 | 412 | H>Y | No | 1000Genomes | |
| ENSVATH00349330 | 416 | R>L | No | 1000Genomes | |
| ENSVATH02201801 | 421 | N>I | No | 1000Genomes | |
| ENSVATH00349331 | 421 | N>K | No | 1000Genomes | |
| ENSVATH00349332 | 422 | K>T | No | 1000Genomes | |
| ENSVATH05912731 | 428 | D>E | No | 1000Genomes | |
| tmp_3_9243097_T_A | 429 | I>N | No | 1000Genomes |
No associated diseases with Q9LSV3
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell surface receptor signaling pathway | The series of molecular signals initiated by activation of a receptor on the surface of a cell. The pathway begins with binding of an extracellular ligand to a cell surface receptor, or for receptors that signal in the absence of a ligand, by ligand-withdrawal or the activity of a constitutively active receptor. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
68 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9Y616 | IRAK3 | Interleukin-1 receptor-associated kinase 3 | Homo sapiens (Human) | PR |
| P53667 | LIMK1 | LIM domain kinase 1 | Homo sapiens (Human) | PR |
| P53671 | LIMK2 | LIM domain kinase 2 | Homo sapiens (Human) | PR |
| P06213 | INSR | Insulin receptor | Homo sapiens (Human) | EV |
| P08069 | IGF1R | Insulin-like growth factor 1 receptor | Homo sapiens (Human) | EV |
| P14616 | INSRR | Insulin receptor-related protein | Homo sapiens (Human) | SS |
| P00533 | EGFR | Epidermal growth factor receptor | Homo sapiens (Human) | EV |
| Q15303 | ERBB4 | Receptor tyrosine-protein kinase erbB-4 | Homo sapiens (Human) | SS |
| P04626 | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | Homo sapiens (Human) | SS |
| P21860 | ERBB3 | Receptor tyrosine-protein kinase erbB-3 | Homo sapiens (Human) | SS |
| Q8K4B2 | Irak3 | Interleukin-1 receptor-associated kinase 3 | Mus musculus (Mouse) | PR |
| Q8CFA1 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Mus musculus (Mouse) | PR |
| O54785 | Limk2 | LIM domain kinase 2 | Mus musculus (Mouse) | PR |
| O08644 | Ephb6 | Ephrin type-B receptor 6 | Mus musculus (Mouse) | PR |
| P54763 | Ephb2 | Ephrin type-B receptor 2 | Mus musculus (Mouse) | SS |
| Q03137 | Epha4 | Ephrin type-A receptor 4 | Mus musculus (Mouse) | SS |
| Q60751 | Igf1r | Insulin-like growth factor 1 receptor | Mus musculus (Mouse) | SS |
| Q61772 | Epha7 | Ephrin type-A receptor 7 | Mus musculus (Mouse) | SS |
| Q8BYG9 | Epha10 | Ephrin type-A receptor 10 | Mus musculus (Mouse) | SS |
| Q8CBF3 | Ephb1 | Ephrin type-B receptor 1 | Mus musculus (Mouse) | SS |
| P54754 | Ephb3 | Ephrin type-B receptor 3 | Mus musculus (Mouse) | SS |
| Q9WTL4 | Insrr | Insulin receptor-related protein | Mus musculus (Mouse) | SS |
| O09127 | Epha8 | Ephrin type-A receptor 8 | Mus musculus (Mouse) | SS |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| P15208 | Insr | Insulin receptor | Mus musculus (Mouse) | SS |
| P54761 | Ephb4 | Ephrin type-B receptor 4 | Mus musculus (Mouse) | PR |
| Q61527 | Erbb4 | Receptor tyrosine-protein kinase erbB-4 | Mus musculus (Mouse) | SS |
| Q61526 | Erbb3 | Receptor tyrosine-protein kinase erbB-3 | Mus musculus (Mouse) | SS |
| Q01279 | Egfr | Epidermal growth factor receptor | Mus musculus (Mouse) | SS |
| Q62413 | Epha6 | Ephrin type-A receptor 6 | Mus musculus (Mouse) | SS |
| P29319 | Epha3 | Ephrin type-A receptor 3 | Mus musculus (Mouse) | SS |
| Q2HWD6 | KIT | Mast/stem cell growth factor receptor Kit | Sus scrofa (Pig) | SS |
| Q4QQS0 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Rattus norvegicus (Rat) | PR |
| O08680 | Epha3 | Ephrin type-A receptor 3 | Rattus norvegicus (Rat) | SS |
| P54757 | Epha5 | Ephrin type-A receptor 5 | Rattus norvegicus (Rat) | SS |
| P54759 | Epha7 | Ephrin type-A receptor 7 | Rattus norvegicus (Rat) | SS |
| P09759 | Ephb1 | Ephrin type-B receptor 1 | Rattus norvegicus (Rat) | SS |
| P0C0K7 | Ephb6 | Ephrin type-B receptor 6 | Rattus norvegicus (Rat) | SS |
| Q9ASQ5 | CRCK3 | Calmodulin-binding receptor-like cytoplasmic kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STF0 | LECRKS3 | Receptor like protein kinase S.3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCZ4 | FER | Receptor-like protein kinase FERONIA | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LYN6 | At3g56050 | Probable inactive receptor-like protein kinase At3g56050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VZJ9 | CRCK2 | Calmodulin-binding receptor-like cytoplasmic kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94C25 | At5g20050 | Probable receptor-like protein kinase At5g20050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FLW0 | At5g24010 | Probable receptor-like protein kinase At5g24010 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LTC0 | PBL19 | Probable serine/threonine-protein kinase PBL19 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FIL7 | CRCK1 | Calmodulin-binding receptor-like cytoplasmic kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7X8C5 | WAKL2 | Wall-associated receptor kinase-like 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9S9M2 | WAKL4 | Wall-associated receptor kinase-like 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LMN7 | WAK5 | Wall-associated receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C9L5 | WAKL9 | Wall-associated receptor kinase-like 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VYA3 | WAKL10 | Wall-associated receptor kinase-like 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M092 | WAKL17 | Wall-associated receptor kinase-like 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8JFR5 | kita | Mast/stem cell growth factor receptor kita | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q9I8N6 | csf1r | Macrophage colony-stimulating factor 1 receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q5GIT4 | kdr | Vascular endothelial growth factor receptor 2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q90Z00 | fgfr1a | Fibroblast growth factor receptor 1-A | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q5MD89 | flt4 | Vascular endothelial growth factor receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q8JG38 | fgfr2 | Fibroblast growth factor receptor 2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q9I8X3 | fgfr3 | Fibroblast growth factor receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKLQHVVYLV | AIFFVVAIFV | IACIEENKYL | VWIMIILANT | TNILSLVRSI | SYIKNIRKHQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KDTKIQRQLF | FEKNGGGMLI | ERLSGAGSSN | IDFKIFTEED | MKEATNGYDV | SRILGQGGQW |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TVYKGILPDN | SIVAIKKTRL | GDNNQVEQFI | NEVLVLSQIN | HRNVVKLLGC | CLETEVPLLV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YEFITGGSLF | DHLHGSMFVS | SLTWEHRLEI | AIEVAGAIAY | LHSGASIPII | HRDIKTENIL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LDENLTAKVA | DFGASKLKPM | DKEQLTTMVQ | GTLGYLDPEY | YTTWLLNEKS | DVYSFGVVLM |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ELISGQKALC | FERPETSKHL | VSYFVLATKE | NRLHEIIDDQ | VLNEENQREI | HEAARVAVEC |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TRLKGEERPR | MIEVAAELET | LRAKTTKHNW | LDQYPEENVH | LLGSNIVSAQ | GHTSSRGYDN |
| 430 | |||||
| NKNVARFDIE | AGR |