Q9LSD2
Gene name |
At3g27040 (MOJ10.11) |
Protein name |
MATH domain and coiled-coil domain-containing protein At3g27040 |
Names |
RTM3-like protein At3g27040 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G27040 |
EC number |
|
Protein Class |
|
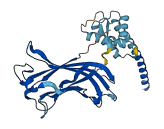
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9LSD2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9LSD2-F1 | Predicted | AlphaFoldDB |
131 variants for Q9LSD2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH11040942 | 2 | G>E | No | 1000Genomes | |
| ENSVATH11040941 | 3 | N>T | No | 1000Genomes | |
| ENSVATH02216171 | 4 | H>R | No | 1000Genomes | |
| tmp_3_9976153_G_A | 4 | H>Y | No | 1000Genomes | |
| tmp_3_9976146_G_A | 6 | A>V | No | 1000Genomes | |
| tmp_3_9976141_TATCAG_CATCAG,T | 8 | K>E | No | 1000Genomes | |
| ENSVATH14028258 | 14 | I>N | No | 1000Genomes | |
| ENSVATH00354446 | 15 | K>M | No | 1000Genomes | |
| ENSVATH11040939 | 21 | G>D | No | 1000Genomes | |
| tmp_3_9976095_C_T | 23 | G>D | No | 1000Genomes | |
| ENSVATH02216170 | 23 | G>S | No | 1000Genomes | |
| ENSVATH00354445 | 24 | S>N | No | 1000Genomes | |
| ENSVATH11040938 | 27 | S>F | No | 1000Genomes | |
| ENSVATH11040938 | 27 | S>Y | No | 1000Genomes | |
| tmp_3_9976068_G_A | 32 | A>V | No | 1000Genomes | |
| ENSVATH02216168 | 33 | G>A | No | 1000Genomes | |
| ENSVATH02216168 | 33 | G>D | No | 1000Genomes | |
| ENSVATH11040924 | 33 | G>S | No | 1000Genomes | |
| ENSVATH11040923 | 34 | R>C | No | 1000Genomes | |
| ENSVATH14028257 | 38 | R>C | No | 1000Genomes | |
| ENSVATH05931968 | 41 | A>V | No | 1000Genomes | |
| ENSVATH11040922 | 43 | P>S | No | 1000Genomes | |
| ENSVATH11040921 | 44 | K>N | No | 1000Genomes | |
| ENSVATH02216163 | 45 | G>E | No | 1000Genomes | |
| ENSVATH05931967 | 45 | G>R | No | 1000Genomes | |
| ENSVATH11040919 | 51 | Y>C | No | 1000Genomes | |
| tmp_3_9975929_A_G | 52 | F>L | No | 1000Genomes | |
| ENSVATH11040918 | 53 | F>S | No | 1000Genomes | |
| ENSVATH11040917 | 54 | L>R | No | 1000Genomes | |
| ENSVATH02216161 | 57 | C>Y | No | 1000Genomes | |
| tmp_3_9975908_G_A | 59 | P>S | No | 1000Genomes | |
| ENSVATH05931966 | 63 | S>P | No | 1000Genomes | |
| ENSVATH11040916 | 64 | L>F | No | 1000Genomes | |
|
ENSVATH02216160 tmp_3_9975880_C_T |
68 | W>* | No | 1000Genomes | |
| ENSVATH02216159 | 70 | R>K | No | 1000Genomes | |
| ENSVATH11040915 | 71 | R>H | No | 1000Genomes | |
| ENSVATH11040903 | 72 | A>D | No | 1000Genomes | |
| ENSVATH11040904 | 72 | A>T | No | 1000Genomes | |
| ENSVATH11040903 | 72 | A>V | No | 1000Genomes | |
| ENSVATH05931964 | 73 | K>N | No | 1000Genomes | |
| ENSVATH02216158 | 74 | V>F | No | 1000Genomes | |
| ENSVATH11040902 | 76 | F>L | No | 1000Genomes | |
| ENSVATH05931962 | 81 | Q>H | No | 1000Genomes | |
| tmp_3_9975835_G_A | 83 | P>L | No | 1000Genomes | |
| ENSVATH14028256 | 85 | G>* | No | 1000Genomes | |
| ENSVATH11040901 | 85 | G>E | No | 1000Genomes | |
| ENSVATH14028256 | 85 | G>R | No | 1000Genomes | |
| tmp_3_9975824_A_C | 87 | S>A | No | 1000Genomes | |
| ENSVATH05931961 | 87 | S>F | No | 1000Genomes | |
| ENSVATH05931960 | 90 | R>Q | No | 1000Genomes | |
| ENSVATH00354444 | 91 | E>K | No | 1000Genomes | |
| ENSVATH02216156 | 92 | A>V | No | 1000Genomes | |
| tmp_3_9975659_A_T | 93 | V>E | No | 1000Genomes | |
| tmp_3_9975660_C_G | 93 | V>L | No | 1000Genomes | |
| tmp_3_9975641_T_G | 99 | K>T | No | 1000Genomes | |
| ENSVATH14028244 | 100 | D>E | No | 1000Genomes | |
| tmp_3_9975639_C_T | 100 | D>N | No | 1000Genomes | |
| tmp_3_9975636_T_A | 101 | T>S | No | 1000Genomes | |
| tmp_3_9975628_A_C | 103 | H>Q | No | 1000Genomes | |
| ENSVATH11040844 | 104 | G>V | No | 1000Genomes | |
| tmp_3_9975621_C_G | 106 | E>Q | No | 1000Genomes | |
| tmp_3_9975608_A_C | 110 | L>R | No | 1000Genomes | |
| tmp_3_9975605_A_C | 111 | L>R | No | 1000Genomes | |
| tmp_3_9975602_G_A | 112 | S>L | No | 1000Genomes | |
| ENSVATH05931952 | 113 | E>D | No | 1000Genomes | |
| ENSVATH05931951 | 114 | I>F | No | 1000Genomes | |
| ENSVATH05931951 | 114 | I>L | No | 1000Genomes | |
| ENSVATH11040843 | 115 | Q>E | No | 1000Genomes | |
| ENSVATH00354439 | 115 | Q>H | No | 1000Genomes | |
| tmp_3_9975593_TG_CG,T | 115 | Q>R | No | 1000Genomes | |
| tmp_3_9975591_T_C | 116 | S>G | No | 1000Genomes | |
| tmp_3_9975590_C_A | 116 | S>I | No | 1000Genomes | |
| ENSVATH11040842 | 118 | D>E | No | 1000Genomes | |
| ENSVATH05931949 | 123 | V>L | No | 1000Genomes | |
| ENSVATH11040839 | 124 | N>D | No | 1000Genomes | |
| tmp_3_9975564_C_T | 125 | G>R | No | 1000Genomes | |
| ENSVATH02216155 | 128 | K>* | No | 1000Genomes | |
| tmp_3_9975548_A_G | 130 | V>A | No | 1000Genomes | |
| ENSVATH05931948 | 130 | V>I | No | 1000Genomes | |
| ENSVATH11040837 | 132 | E>K | No | 1000Genomes | |
| tmp_3_9975540_C_T | 133 | V>I | No | 1000Genomes | |
| ENSVATH05931947 | 136 | L>F | No | 1000Genomes | |
| ENSVATH14028243 | 144 | V>L | No | 1000Genomes | |
| ENSVATH14028242 | 146 | E>D | No | 1000Genomes | |
| ENSVATH11040836 | 146 | E>K | No | 1000Genomes | |
| tmp_3_9975498_C_A | 147 | E>* | No | 1000Genomes | |
| ENSVATH11040835 | 150 | R>S | No | 1000Genomes | |
| ENSVATH11040823 | 156 | F>L | No | 1000Genomes | |
| ENSVATH11040824 | 156 | F>Y | No | 1000Genomes | |
| ENSVATH11040822 | 157 | Q>E | No | 1000Genomes | |
| ENSVATH11040821 | 158 | V>I | No | 1000Genomes | |
| ENSVATH11040818 | 159 | P>L | No | 1000Genomes | |
| ENSVATH14028241 | 160 | A>D | No | 1000Genomes | |
| ENSVATH11040817 | 160 | A>T | No | 1000Genomes | |
| tmp_3_9975455_G_A | 161 | S>L | No | 1000Genomes | |
| ENSVATH05931945 | 162 | Q>H | No | 1000Genomes | |
| tmp_3_9975234_G_A | 165 | S>F | No | 1000Genomes | |
| tmp_3_9975230_C_T | 166 | M>I | No | 1000Genomes | |
| tmp_3_9975220_A_T | 170 | F>I | No | 1000Genomes | |
| ENSVATH05931934 | 172 | K>N | No | 1000Genomes | |
| tmp_3_9975214_T_G | 172 | K>Q | No | 1000Genomes | |
| tmp_3_9975213_T_C | 172 | K>R | No | 1000Genomes | |
| tmp_3_9975207_C_T | 174 | R>Q | No | 1000Genomes | |
| tmp_3_9975205_C_T | 175 | G>S | No | 1000Genomes | |
| ENSVATH11040768 | 182 | P>S | No | 1000Genomes | |
| ENSVATH11040767 | 187 | L>Q | No | 1000Genomes | |
| ENSVATH11040766 | 192 | L>M | No | 1000Genomes | |
| tmp_3_9975141_A_G | 196 | L>P | No | 1000Genomes | |
| tmp_3_9975134_C_T | 198 | M>I | No | 1000Genomes | |
| tmp_3_9975136_T_G | 198 | M>L | No | 1000Genomes | |
| ENSVATH11040765 | 200 | E>K | No | 1000Genomes | |
| ENSVATH05931931 | 203 | C>S | No | 1000Genomes | |
| ENSVATH00354435 | 205 | F>S | No | 1000Genomes | |
| ENSVATH05931930 | 206 | P>A | No | 1000Genomes | |
| tmp_3_9975111_G_A | 206 | P>L | No | 1000Genomes | |
| ENSVATH05931929 | 207 | E>K | No | 1000Genomes | |
| tmp_3_9975106_C_A | 208 | E>* | No | 1000Genomes | |
| ENSVATH05931926 | 210 | S>T | No | 1000Genomes | |
| ENSVATH05931925 | 210 | S>Y | No | 1000Genomes | |
| tmp_3_9975096_C_T | 211 | S>N | No | 1000Genomes | |
| ENSVATH14028236 | 215 | A>V | No | 1000Genomes | |
| ENSVATH05931924 | 220 | A>T | No | 1000Genomes | |
| tmp_3_9975058_C_T | 224 | V>I | No | 1000Genomes | |
| ENSVATH05931923 | 228 | G>E | No | 1000Genomes | |
| ENSVATH00354434 | 243 | K>N | No | 1000Genomes | |
| ENSVATH02216148 | 248 | E>K | No | 1000Genomes | |
| ENSVATH14028235 | 253 | L>* | No | 1000Genomes | |
| ENSVATH05931919 | 261 | A>T | No | 1000Genomes | |
| ENSVATH00354433 | 268 | D>E | No | 1000Genomes | |
| ENSVATH11040713 | 270 | L>F | No | 1000Genomes | |
| ENSVATH14028194 | 271 | R>* | No | 1000Genomes |
No associated diseases with Q9LSD2
1 regional properties for Q9LSD2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Ubiquitin-conjugating enzyme E2 | 1 - 154 | IPR000608 |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q6IQ16 | SPOPL | Speckle-type POZ protein-like | Homo sapiens (Human) | PR |
| Q6YCH2 | Tdpoz4 | TD and POZ domain-containing protein 4 | Mus musculus (Mouse) | PR |
| Q2M2N2 | Spopl | Speckle-type POZ protein-like | Mus musculus (Mouse) | PR |
| P34371 | bath-42 | BTB and MATH domain-containing protein 42 | Caenorhabditis elegans | PR |
| F4IN32 | At2g42460 | MATH domain and coiled-coil domain-containing protein At2g42460 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4J4A0 | At3g44790 | MATH domain and coiled-coil domain-containing protein At3g44790 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4J4A1 | At3g44800 | MATH domain and coiled-coil domain-containing protein At3g44800 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0DKG7 | At2g42480 | MATH domain and coiled-coil domain-containing protein At2g42480 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2I2 | At3g58340 | MATH domain and coiled-coil domain-containing protein At3g58340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2J0 | At3g58260 | MATH domain and coiled-coil domain-containing protein At3g58260 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2J5 | At3g58210 | MATH domain and coiled-coil domain-containing protein At3g58210 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZUA7 | At2g01790 | MATH domain and coiled-coil domain-containing protein At2g01790 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0DKG4 | At2g42465 | MATH domain and coiled-coil domain-containing protein At2g42465 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SHT3 | At2g05410 | MATH domain and coiled-coil domain-containing protein At2g05410 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4J4P8 | At3g58220 | MATH domain and coiled-coil domain-containing protein At3g58220 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGNHQADKKF | TWVIKNYNSL | GSGSVYSDTF | KAGRCKWRLL | AFPKGNNIYD | YFFLYICVPN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SESLPSGWRR | RAKVSFTMVN | QIPGGLSQQR | EAVYWFDEKD | TTHGFESMFL | LSEIQSSDKG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| FLVNGEVKIV | AEVDVLEVIG | ELDVPEEPER | IDINGFQVPA | SQVESMNSLF | EKYRGFASKI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FPKNQHLRKT | FLDVVLSMTE | ILCKFPEELS | SGDLAEAYSA | LRFVTKAGFK | LDWLEKKLKE |
| 250 | 260 | 270 | |||
| TGKSRLQEIE | EDLKDLKVKC | ADMDALLDFL | R |