Q9LNK1
Gene name |
RABA3 (At1g01200, F6F3.1) |
Protein name |
Ras-related protein RABA3 |
Names |
AtRABA3 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G01200 |
EC number |
|
Protein Class |
|
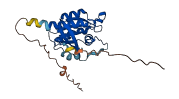
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9LNK1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9LNK1-F1 | Predicted | AlphaFoldDB |
18 variants for Q9LNK1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_1_88112_T_C | 12 | N>D | No | 1000Genomes | |
| ENSVATH04501294 | 16 | V>A | No | 1000Genomes | |
| ENSVATH07500213 | 20 | T>K | No | 1000Genomes | |
| ENSVATH00000341 | 54 | C>Y | No | 1000Genomes | |
| ENSVATH10440379 | 66 | Q>R | No | 1000Genomes | |
| tmp_1_87154_G_A | 92 | A>V | No | 1000Genomes | |
| ENSVATH00000337 | 148 | G>D | No | 1000Genomes | |
| tmp_1_86980_C_A | 150 | R>L | No | 1000Genomes | |
| ENSVATH01000963 | 158 | V>L | No | 1000Genomes | |
| ENSVATH13847966 | 161 | A>S | No | 1000Genomes | |
| ENSVATH13847965 | 162 | E>K | No | 1000Genomes | |
| tmp_1_86939_G_C | 164 | Q>E | No | 1000Genomes | |
| ENSVATH04501279 | 165 | R>Q | No | 1000Genomes | |
| ENSVATH04501277 | 185 | R>H | No | 1000Genomes | |
| tmp_1_86867_C_T | 188 | E>K | No | 1000Genomes | |
| tmp_1_86851_C_T | 193 | R>H | No | 1000Genomes | |
| ENSVATH04501276 | 200 | A>G | No | 1000Genomes | |
| ENSVATH13847964 | 200 | A>S | No | 1000Genomes |
No associated diseases with Q9LNK1
1 regional properties for Q9LNK1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 28 - 183 | IPR005225 |
Functions
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell plate | The nascent cell membrane and cell wall structure that forms between two daughter nuclei near the center of a dividing plant cell. It develops at the equitorial region of the phragmoplast. It grows outwards to join with the lateral walls and form two daughter cells. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| endosome membrane | The lipid bilayer surrounding an endosome. |
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell wall biogenesis | A cellular process that results in the biosynthesis of constituent macromolecules, assembly, and arrangement of constituent parts of a cell wall. Includes biosynthesis of constituent macromolecules, such as proteins and polysaccharides, and those macromolecular modifications that are involved in synthesis or assembly of the cellular component. A cell wall is the rigid or semi-rigid envelope lying outside the cell membrane of plant, fungal and most prokaryotic cells, maintaining their shape and protecting them from osmotic lysis. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
23 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3MHP2 | RAB11B | Ras-related protein Rab-11B | Bos taurus (Bovine) | PR |
| Q2TA29 | RAB11A | Ras-related protein Rab-11A | Bos taurus (Bovine) | PR |
| Q5ZJN2 | RAB11A | Ras-related protein Rab-11A | Gallus gallus (Chicken) | PR |
| Q5ZHV1 | RAB33B | Ras-related protein Rab-33B | Gallus gallus (Chicken) | PR |
| P62490 | RAB11A | Ras-related protein Rab-11A | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| P62491 | RAB11A | Ras-related protein Rab-11A | Homo sapiens (Human) | PR |
| Q15907 | RAB11B | Ras-related protein Rab-11B | Homo sapiens (Human) | PR |
| Q96AX2 | RAB37 | Ras-related protein Rab-37 | Homo sapiens (Human) | PR |
| Q9H082 | RAB33B | Ras-related protein Rab-33B | Homo sapiens (Human) | PR |
| P46638 | Rab11b | Ras-related protein Rab-11B | Mus musculus (Mouse) | PR |
| P62492 | Rab11a | Ras-related protein Rab-11A | Mus musculus (Mouse) | PR |
| Q9JKM7 | Rab37 | Ras-related protein Rab-37 | Mus musculus (Mouse) | PR |
| O35963 | Rab33b | Ras-related protein Rab-33B | Mus musculus (Mouse) | PR |
| Q52NJ1 | RAB11A | Ras-related protein Rab-11A | Sus scrofa (Pig) | PR |
| O35509 | Rab11b | Ras-related protein Rab-11B | Rattus norvegicus (Rat) | PR |
| P62494 | Rab11a | Ras-related protein Rab-11A | Rattus norvegicus (Rat) | PR |
| Q20365 | rab-33 | Ras-related protein Rab-33 | Caenorhabditis elegans | PR |
| O04486 | RABA2A | Ras-related protein RABA2a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O49513 | RABA1E | Ras-related protein RABA1e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FE79 | RABA4C | Ras-related protein RABA4c | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LH50 | RABA4D | Ras-related protein RABA4d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LNW1 | RABA2B | Ras-related protein RABA2b | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJZ5 | RABA4E | Putative Ras-related protein RABA4e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNEEMSGESP | ENNKHVKKPT | MPEKIDYVFK | VVVIGDSAVG | KTQLLSRFTH | NEFCYDSKST |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IGVEFQTRTI | TLRGKLVKAQ | IWDTAGQERY | RAVTSAYYRG | ALGAMVVYDI | TKRLSFDHVA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RWVEELRAHA | DDSAVIMLVG | NKADLSVGKR | AVPTEDAVEF | AETQRLFFSE | VSALSGGNVD |
| 190 | 200 | 210 | 220 | 230 | |
| EAFFRLLEEI | FSRVVVSRKA | MESDGGATVK | LDGSRIDVIS | GSDLETSNIK | EQASCSC |