Q9LH02
Gene name |
FH17 (At3g32400, F1D9.13) |
Protein name |
Formin-like protein 17 |
Names |
AtFH17 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G32400 |
EC number |
|
Protein Class |
|
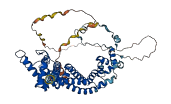
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9LH02
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9LH02-F1 | Predicted | AlphaFoldDB |
150 variants for Q9LH02
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_3_13360623_A_C | 3 | I>S | No | 1000Genomes | |
| ENSVATH06076033 | 4 | R>K | No | 1000Genomes | |
| ENSVATH02323891 | 7 | I>V | No | 1000Genomes | |
| ENSVATH00379206 | 8 | D>H | No | 1000Genomes | |
| ENSVATH00379206 | 8 | D>N | No | 1000Genomes | |
| tmp_3_13360603_T_C | 10 | T>A | No | 1000Genomes | |
| ENSVATH02323890 | 10 | T>R | No | 1000Genomes | |
| ENSVATH08053843 | 11 | E>K | No | 1000Genomes | |
| ENSVATH00379203 | 13 | S>N | No | 1000Genomes | |
| ENSVATH11770326 | 16 | A>E | No | 1000Genomes | |
| ENSVATH08053819 | 17 | T>I | No | 1000Genomes | |
| ENSVATH02323849 | 18 | E>* | No | 1000Genomes | |
| ENSVATH06076016 | 18 | E>D | No | 1000Genomes | |
| ENSVATH02323849 | 18 | E>K | No | 1000Genomes | |
| ENSVATH02323849 | 18 | E>Q | No | 1000Genomes | |
| ENSVATH08053818 | 19 | G>R | No | 1000Genomes | |
| ENSVATH11770325 | 23 | P>A | No | 1000Genomes | |
| ENSVATH11770325 | 23 | P>S | No | 1000Genomes | |
| ENSVATH02323846 | 34 | S>F | No | 1000Genomes | |
| ENSVATH02323844 | 36 | L>F | No | 1000Genomes | |
| ENSVATH06076015 | 37 | S>* | No | 1000Genomes | |
| ENSVATH06076015 | 37 | S>L | No | 1000Genomes | |
| ENSVATH02323842 | 42 | P>L | No | 1000Genomes | |
| ENSVATH14202640 | 45 | L>S | No | 1000Genomes | |
| tmp_3_13360000_G_C | 46 | P>R | No | 1000Genomes | |
| ENSVATH11770301 | 50 | L>F | No | 1000Genomes | |
| tmp_3_13359961_G_A | 59 | P>L | No | 1000Genomes | |
| tmp_3_13359958_G_A | 60 | P>L | No | 1000Genomes | |
| ENSVATH14202638 | 66 | S>C | No | 1000Genomes | |
| tmp_3_13359936_A_T | 67 | N>K | No | 1000Genomes | |
| tmp_3_13359937_T_C | 67 | N>S | No | 1000Genomes | |
| ENSVATH06076014 | 69 | Q>L | No | 1000Genomes | |
| ENSVATH14202637 | 71 | G>E | No | 1000Genomes | |
| ENSVATH14202635 | 72 | A>S | No | 1000Genomes | |
| ENSVATH14202632 | 82 | P>L | No | 1000Genomes | |
| tmp_3_13359754_T_C | 93 | K>E | No | 1000Genomes | |
| ENSVATH02323830 | 109 | Q>H | No | 1000Genomes | |
| tmp_3_13359687_T_C | 115 | E>G | No | 1000Genomes | |
| ENSVATH02323829 | 117 | Q>* | No | 1000Genomes | |
| tmp_3_13359563_C_T | 127 | D>N | No | 1000Genomes | |
| tmp_3_13359551_A_G | 131 | S>P | No | 1000Genomes | |
| tmp_3_13359548_C_T | 132 | E>K | No | 1000Genomes | |
| ENSVATH02323824 | 133 | I>L | No | 1000Genomes | |
| ENSVATH02323823 | 139 | A>V | No | 1000Genomes | |
| ENSVATH02323822 | 144 | S>F | No | 1000Genomes | |
| ENSVATH02323821 | 145 | N>D | No | 1000Genomes | |
| tmp_3_13359504_A_T | 146 | S>R | No | 1000Genomes | |
| ENSVATH02323820 | 150 | G>C | No | 1000Genomes | |
| ENSVATH11770241 | 151 | G>E | No | 1000Genomes | |
| ENSVATH11770240 | 155 | R>* | No | 1000Genomes | |
| ENSVATH08053808 | 156 | R>C | No | 1000Genomes | |
| tmp_3_13359475_C_T | 156 | R>H | No | 1000Genomes | |
| ENSVATH02323818 | 158 | R>G | No | 1000Genomes | |
| tmp_3_13359463_T_G | 160 | K>T | No | 1000Genomes | |
| ENSVATH11770238 | 163 | K>R | No | 1000Genomes | |
| tmp_3_13359449_G_A | 165 | Q>* | No | 1000Genomes | |
| ENSVATH02323806 | 170 | K>R | No | 1000Genomes | |
| ENSVATH02323805 | 175 | C>S | No | 1000Genomes | |
| tmp_3_13359264_T_C | 181 | K>E | No | 1000Genomes | |
| ENSVATH06075996 | 185 | P>L | No | 1000Genomes | |
| ENSVATH06075996 | 185 | P>R | No | 1000Genomes | |
| ENSVATH02323804 | 187 | P>H | No | 1000Genomes | |
| ENSVATH02323804 | 187 | P>L | No | 1000Genomes | |
| tmp_3_13359032_T_C | 207 | D>G | No | 1000Genomes | |
| ENSVATH02323795 | 217 | E>D | No | 1000Genomes | |
| ENSVATH11770176 | 218 | E>K | No | 1000Genomes | |
| ENSVATH02323787 | 226 | I>T | No | 1000Genomes | |
| tmp_3_13358837_C_T | 227 | G>E | No | 1000Genomes | |
| ENSVATH02323786 | 227 | G>R | No | 1000Genomes | |
| ENSVATH06075983 | 234 | R>Q | No | 1000Genomes | |
| ENSVATH06075981 | 235 | C>* | No | 1000Genomes | |
| ENSVATH06075982 | 235 | C>G | No | 1000Genomes | |
| tmp_3_13358674_C_G | 245 | V>L | No | 1000Genomes | |
| ENSVATH06075972 | 264 | Q>* | No | 1000Genomes | |
| ENSVATH06075972 | 264 | Q>E | No | 1000Genomes | |
| ENSVATH02323773 | 270 | R>T | No | 1000Genomes | |
| tmp_3_13358502_G_T | 274 | T>N | No | 1000Genomes | |
| ENSVATH06075970 | 276 | H>N | No | 1000Genomes | |
| ENSVATH02323772 | 279 | T>A | No | 1000Genomes | |
| ENSVATH06075966 | 282 | V>I | No | 1000Genomes | |
| ENSVATH02323767 | 284 | G>R | No | 1000Genomes | |
| ENSVATH02323766 | 286 | T>A | No | 1000Genomes | |
| tmp_3_13358369_T_C | 290 | R>G | No | 1000Genomes | |
| tmp_3_13358368_C_T | 290 | R>K | No | 1000Genomes | |
| tmp_3_13358361_C_G | 292 | M>I | No | 1000Genomes | |
| tmp_3_13358356_G_T | 294 | T>N | No | 1000Genomes | |
| tmp_3_13358342_C_A | 299 | G>C | No | 1000Genomes | |
| tmp_3_13358326_T_C | 304 | H>R | No | 1000Genomes | |
| ENSVATH11770075 | 305 | G>E | No | 1000Genomes | |
| ENSVATH11770044 | 306 | T>I | No | 1000Genomes | |
| tmp_3_13358317_G_C | 307 | A>G | No | 1000Genomes | |
| ENSVATH02323764 | 307 | A>T | No | 1000Genomes | |
| ENSVATH02323756 | 313 | G>R | No | 1000Genomes | |
| tmp_3_13358170_C_A | 313 | G>V | No | 1000Genomes | |
| tmp_3_13358165_G_A | 315 | H>Y | No | 1000Genomes | |
| tmp_3_13358162_G_T | 316 | L>M | No | 1000Genomes | |
| tmp_3_13358140_G_C | 323 | T>S | No | 1000Genomes | |
| ENSVATH02323755 | 324 | D>N | No | 1000Genomes | |
| ENSVATH02323755 | 324 | D>Y | No | 1000Genomes | |
| tmp_3_13358134_G_A | 325 | T>I | No | 1000Genomes | |
| tmp_3_13358126_G_C | 328 | R>G | No | 1000Genomes | |
| ENSVATH02323754 | 328 | R>H | No | 1000Genomes | |
| ENSVATH02323754 | 328 | R>L | No | 1000Genomes | |
| ENSVATH06075957 | 329 | N>K | No | 1000Genomes | |
| ENSVATH06075956 | 330 | S>R | No | 1000Genomes | |
| ENSVATH06075955 | 331 | K>T | No | 1000Genomes | |
| tmp_3_13358107_A_T | 334 | L>Q | No | 1000Genomes | |
| ENSVATH14202553 | 335 | M>I | No | 1000Genomes | |
| ENSVATH14202552 | 336 | H>D | No | 1000Genomes | |
| ENSVATH14202551 | 336 | H>Q | No | 1000Genomes | |
| tmp_3_13358095_A_C | 338 | L>R | No | 1000Genomes | |
| ENSVATH02323745 | 341 | V>L | No | 1000Genomes | |
| tmp_3_13357913_C_T | 343 | A>T | No | 1000Genomes | |
| ENSVATH06075947 | 343 | A>V | No | 1000Genomes | |
| tmp_3_13357904_G_A | 346 | L>F | No | 1000Genomes | |
| tmp_3_13357900_G_T | 347 | P>Q | No | 1000Genomes | |
| ENSVATH02323744 | 348 | G>E | No | 1000Genomes | |
| tmp_3_13357898_C_A | 348 | G>W | No | 1000Genomes | |
| ENSVATH06075946 | 355 | D>N | No | 1000Genomes | |
| ENSVATH02323743 | 356 | M>L | No | 1000Genomes | |
| ENSVATH11770038 | 356 | M>T | No | 1000Genomes | |
| ENSVATH11770037 | 358 | S>G | No | 1000Genomes | |
| ENSVATH06075943 | 358 | S>R | No | 1000Genomes | |
| ENSVATH06075944 | 358 | S>T | No | 1000Genomes | |
| ENSVATH00379191 | 364 | N>K | No | 1000Genomes | |
| ENSVATH06075940 | 365 | I>F | No | 1000Genomes | |
| tmp_3_13357730_G_T,A | 371 | A>E | No | 1000Genomes | |
| tmp_3_13357730_G_T,A | 371 | A>V | No | 1000Genomes | |
| ENSVATH11769999 | 374 | M>I | No | 1000Genomes | |
| ENSVATH11770000 | 374 | M>K | No | 1000Genomes | |
| ENSVATH11769998 | 376 | A>P | No | 1000Genomes | |
| ENSVATH02323740 | 381 | L>F | No | 1000Genomes | |
| ENSVATH11769997 | 382 | E>D | No | 1000Genomes | |
| tmp_3_13357670_G_A | 391 | S>F | No | 1000Genomes | |
| tmp_3_13357662_C_T | 394 | D>N | No | 1000Genomes | |
| ENSVATH02323739 | 395 | C>G | No | 1000Genomes | |
| tmp_3_13357656_GA_CA,G | 396 | Q>E | No | 1000Genomes | |
| ENSVATH02323738 | 402 | H>R | No | 1000Genomes | |
| ENSVATH02323737 | 403 | M>I | No | 1000Genomes | |
| ENSVATH06075935 | 411 | V>F | No | 1000Genomes | |
| ENSVATH11769944 | 437 | F>L | No | 1000Genomes | |
| ENSVATH02323731 | 438 | G>E | No | 1000Genomes | |
| tmp_3_13357104_A_G | 460 | F>S | No | 1000Genomes | |
| ENSVATH11769896 | 463 | S>A | No | 1000Genomes | |
| tmp_3_13357036_C_T | 483 | E>K | No | 1000Genomes | |
| tmp_3_13357030_C_T | 485 | E>K | No | 1000Genomes | |
| ENSVATH02323723 | 487 | L>H | No | 1000Genomes | |
| ENSVATH02323722 | 488 | K>N | No | 1000Genomes | |
| ENSVATH02323721 | 490 | G>D | No | 1000Genomes | |
| ENSVATH14202514 | 491 | V>F | No | 1000Genomes |
No associated diseases with Q9LH02
1 regional properties for Q9LH02
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Formin, FH2 domain | 18 - 495 | IPR015425 |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A0A1D5P556 | DAAM2 | Disheveled-associated activator of morphogenesis 2 | Gallus gallus (Chicken) | SS |
| O95466 | FMNL1 | Formin-like protein 1 | Homo sapiens (Human) | SS |
| Q27J81 | INF2 | Inverted formin-2 | Homo sapiens (Human) | EV |
| Q9Y4D1 | DAAM1 | Disheveled-associated activator of morphogenesis 1 | Homo sapiens (Human) | EV |
| Q86T65 | DAAM2 | Disheveled-associated activator of morphogenesis 2 | Homo sapiens (Human) | SS |
| Q80U19 | Daam2 | Disheveled-associated activator of morphogenesis 2 | Mus musculus (Mouse) | SS |
| Q8BPM0 | Daam1 | Disheveled-associated activator of morphogenesis 1 | Mus musculus (Mouse) | PR |
| Q0GNC1 | Inf2 | Inverted formin-2 | Mus musculus (Mouse) | SS |
| Q0QWG9 | Grid2ip | Delphilin | Mus musculus (Mouse) | PR |
| Q9C7S1 | FH12 | Formin-like protein 12 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0C5K4 | FH21A | Putative formin-like protein 21a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FF14 | FH19 | Formin-like protein 19 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDIRELIDIT | ELSSIATEGP | PPPPPPPLLQ | PHHSALSSSP | LPPPLPPKKL | LATTNTPPPP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PPPLHSNSQM | GAPTSSLVLK | SPHNLKGQGQ | TRKANLKPYH | WLKLTRAVQG | SLWAEAQKSD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EAATAPDFDI | SEIEKLFSAV | NLSSNSENNG | GKSGRRARPK | VEKVQLIELK | RAYNCEIMLS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KVKIPLPDLM | SSVLALDESV | IDVDQVDNLI | KFCPTKEEAE | LLKGFIGNKE | TLGRCEQFFL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ELLKVPRVET | KLRVFSFKIQ | FHSQVTDLRR | GLNTIHSATN | EVRGSTKLKR | IMQTILSLGN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ALNHGTARGS | AIGFHLDSLL | KLTDTRSRNS | KMTLMHYLCK | VLAEKLPGLL | NFPKDMVSLE |
| 370 | 380 | 390 | 400 | 410 | 420 |
| AATNIQLKYL | AEEMQATSKG | LEKVVQEFTA | SETDCQISKH | FHMNLKEFLS | VAEGEVRSLA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SLYSTVGGSA | DALALYFGED | PARVPFEQVV | STLQNFVRIF | VRSHEENCKQ | VEFEKKRAQK |
| 490 | |||||
| EAENEKLKKG | VYNEN |