Q9LDR2
Gene name |
CNGC19 (CNBT2, At3g17690, MKP6.27) |
Protein name |
Putative cyclic nucleotide-gated ion channel 19 |
Names |
Cyclic nucleotide-binding transporter 2 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G17690 |
EC number |
|
Protein Class |
|
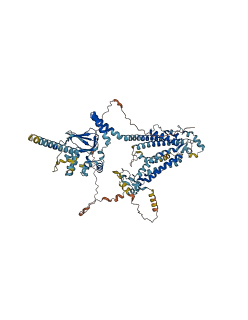
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9LDR2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9LDR2-F1 | Predicted | AlphaFoldDB |
37 variants for Q9LDR2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH00329722 | 8 | T>S | No | 1000Genomes | |
| ENSVATH05852289 | 12 | R>C | No | 1000Genomes | |
| ENSVATH05852290 | 13 | S>R | No | 1000Genomes | |
| ENSVATH00329723 | 32 | T>I | No | 1000Genomes | |
| tmp_3_6045481_G_A | 34 | G>S | No | 1000Genomes | |
| ENSVATH05852291 | 41 | T>S | No | 1000Genomes | |
| ENSVATH00329724 | 43 | R>K | No | 1000Genomes | |
| tmp_3_6045511_A_G | 44 | I>V | No | 1000Genomes | |
| tmp_3_6045533_T_A | 51 | M>K | No | 1000Genomes | |
| tmp_3_6045547_C_T | 56 | H>Y | No | 1000Genomes | |
| tmp_3_6045578_C_A | 66 | S>Y | No | 1000Genomes | |
| ENSVATH05852292 | 109 | T>S | No | 1000Genomes | |
| ENSVATH13937586 | 113 | S>P | No | 1000Genomes | |
| tmp_3_6045752_C_A | 124 | T>N | No | 1000Genomes | |
| ENSVATH05852293 | 125 | S>Y | No | 1000Genomes | |
| tmp_3_6045884_G_A | 138 | G>D | No | 1000Genomes | |
| tmp_3_6045902_C_T | 144 | A>V | No | 1000Genomes | |
| ENSVATH02157178 | 153 | R>S | No | 1000Genomes | |
| tmp_3_6046323_C_G | 235 | A>G | No | 1000Genomes | |
| ENSVATH10746059 | 254 | A>P | No | 1000Genomes | |
| tmp_3_6046402_G_T | 261 | K>N | No | 1000Genomes | |
| tmp_3_6046407_T_C | 263 | L>P | No | 1000Genomes | |
| ENSVATH05852300 | 300 | T>M | No | 1000Genomes | |
| tmp_3_6046542_A_C | 308 | K>T | No | 1000Genomes | |
| tmp_3_6046808_G_C | 367 | W>S | No | 1000Genomes | |
| tmp_3_6046832_G_T | 375 | R>I | No | 1000Genomes | |
| tmp_3_6046833_A_C | 375 | R>S | No | 1000Genomes | |
| tmp_3_6046904_G_A | 399 | S>N | No | 1000Genomes | |
| tmp_3_6046916_G_A | 403 | C>Y | No | 1000Genomes | |
| tmp_3_6047000_G_A | 431 | S>N | No | 1000Genomes | |
| ENSVATH05852303 | 435 | Y>F | No | 1000Genomes | |
| tmp_3_6047523_C_A | 525 | A>D | No | 1000Genomes | |
| tmp_3_6047773_A_G | 572 | I>V | No | 1000Genomes | |
| tmp_3_6047846_A_G | 596 | K>R | No | 1000Genomes | |
| tmp_3_6047962_A_C | 635 | I>L | No | 1000Genomes | |
| tmp_3_6048083_C_T | 645 | P>L | No | 1000Genomes | |
| tmp_3_6048326_A_G | 726 | N>S | No | 1000Genomes |
No associated diseases with Q9LDR2
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| cAMP binding | Binding to cAMP, the nucleotide cyclic AMP (adenosine 3',5'-cyclophosphate). |
| cGMP binding | Binding to cGMP, the nucleotide cyclic GMP (guanosine 3',5'-cyclophosphate). |
| ligand-gated calcium channel activity | Enables the transmembrane transfer of a calcium ions by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| response to herbivore | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a herbivore. |
31 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q03041 | CNGA2 | Cyclic nucleotide-gated olfactory channel | Bos taurus (Bovine) | PR |
| Q00194 | CNGA1 | cGMP-gated cation channel alpha-1 | Bos taurus (Bovine) | PR |
| Q29441 | CNGA3 | Cyclic nucleotide-gated cation channel alpha-3 | Bos taurus (Bovine) | PR |
| Q90980 | Cyclic nucleotide-gated channel rod photoreceptor subunit alpha | Gallus gallus (Chicken) | PR | |
| Q28279 | CNGA1 | cGMP-gated cation channel alpha-1 | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| Q9UL51 | HCN2 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | Homo sapiens (Human) | SS |
| Q9Y3Q4 | HCN4 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | Homo sapiens (Human) | EV |
| O60741 | HCN1 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | Homo sapiens (Human) | SS |
| Q9P1Z3 | HCN3 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | Homo sapiens (Human) | SS |
| Q16281 | CNGA3 | Cyclic nucleotide-gated cation channel alpha-3 | Homo sapiens (Human) | PR |
| P29973 | CNGA1 | cGMP-gated cation channel alpha-1 | Homo sapiens (Human) | PR |
| Q16280 | CNGA2 | Cyclic nucleotide-gated olfactory channel | Homo sapiens (Human) | PR |
| O88704 | Hcn1 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | Mus musculus (Mouse) | SS |
| O88705 | Hcn3 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | Mus musculus (Mouse) | SS |
| O70507 | Hcn4 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | Mus musculus (Mouse) | SS |
| O88703 | Hcn2 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | Mus musculus (Mouse) | EV |
| Q9JJZ8 | Cnga3 | Cyclic nucleotide-gated cation channel alpha-3 | Mus musculus (Mouse) | PR |
| P29974 | Cnga1 | cGMP-gated cation channel alpha-1 | Mus musculus (Mouse) | PR |
| Q62398 | Cnga2 | Cyclic nucleotide-gated olfactory channel | Mus musculus (Mouse) | PR |
| Q9JKA9 | Hcn2 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | Rattus norvegicus (Rat) | SS |
| Q9JKA7 | Hcn4 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | Rattus norvegicus (Rat) | SS |
| Q9JKB0 | Hcn1 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | Rattus norvegicus (Rat) | SS |
| Q9JKA8 | Hcn3 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | Rattus norvegicus (Rat) | SS |
| Q62927 | Cnga1 | cGMP-gated cation channel alpha-1 | Rattus norvegicus (Rat) | PR |
| Q00195 | Cnga2 | Cyclic nucleotide-gated olfactory channel | Rattus norvegicus (Rat) | PR |
| A2ZX97 | Os01g0718700 | Potassium channel KAT6 | Oryza sativa subsp japonica (Rice) | PR |
| Q6K3T2 | Os02g0245800 | Potassium channel KAT1 | Oryza sativa subsp japonica (Rice) | PR |
| P92960 | ATHB-4 | Potassium channel KAT3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39128 | KAT1 | Potassium channel KAT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SKD7 | CNGC3 | Probable cyclic nucleotide-gated ion channel 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SU64 | CNGC16 | Probable cyclic nucleotide-gated ion channel 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAHTRTFTSR | NRSVSLSNPS | FSIDGFDNST | VTLGYTGPLR | TQRIRPPLVQ | MSGPIHSTRR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TEPLFSPSPQ | ESPDSSSTVD | VPPEDDFVFK | NANLLRSGQL | GMCNDPYCTT | CPSYYNRQAA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QLHTSRVSAS | RFRTVLYGDA | RGWAKRFASS | VRRCLPGIMN | PHSKFVQVWT | RVLAFSSLVA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IFIDPLFFFL | LLIQQDNKCI | AIDWRATKVL | VSLRSITDLI | FFINILLQFR | LAYVAPESRI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VGAGQLVDHP | RKIARHYFRG | KFLLDMFIVF | PIPQIMILRI | IPLHLGTRRE | ESEKQILRAT |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VLFQYIPKLY | RLLPLLAGQT | STGFIFESAW | ANFVINLLTF | MLAGHAVGSC | WYLSALQRVK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| KCMLNAWNIS | ADERRNLIDC | ARGSYASKSQ | RDLWRDNASV | NACFQENGYT | YGIYLKAVNL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TNESSFFTRF | SYSLYWGFQQ | ISTLAGNLSP | SYSVGEVFFT | MGIIGLGLLL | FARLIGNMHN |
| 490 | 500 | 510 | 520 | 530 | 540 |
| FLQSLDRRRM | EMMLRKRDVE | QWMSHRRLPE | DIRKRVREVE | RYTWAATRGV | NEELLFENMP |
| 550 | 560 | 570 | 580 | 590 | 600 |
| DDLQRDIRRH | LFKFLKKVRI | FSLMDESVLD | SIRERLKQRT | YIRSSTVLHH | RGLVEKMVFI |
| 610 | 620 | 630 | 640 | 650 | 660 |
| VRGEMESIGE | DGSVLPLSEG | DVCGEELLTW | CLSSINPDGT | RIKMPPKGLV | SNRNVRCVTN |
| 670 | 680 | 690 | 700 | 710 | 720 |
| VEAFSLSVAD | LEDVTSLFSR | FLRSHRVQGA | IRYESPYWRL | RAAMQIQVAW | RYRKRQLQRL |
| NTAHSNSNR |