Q9JMK2
Gene name |
Csnk1e |
Protein name |
Casein kinase I isoform epsilon |
Names |
CKI-epsilon, CKIe |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:27373 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
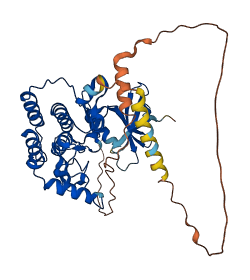
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
9-287 (Protein kinase domain) |
Relief mechanism |
Cleavage, PTM |
Assay |
|
Accessory elements
148-178 (Activation loop from InterPro)
Target domain |
9-287 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9JMK2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9JMK2-F1 | Predicted | AlphaFoldDB |
1 variants for Q9JMK2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs238047958 | 370 | A>V | No | Ensembl |
No associated diseases with Q9JMK2
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| growth cone | The migrating motile tip of a growing neuron projection, where actin accumulates, and the actin cytoskeleton is the most dynamic. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| ribonucleoprotein complex | A macromolecular complex that contains both RNA and protein molecules. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to nerve growth factor stimulus | A process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nerve growth factor stimulus. |
| circadian behavior | The specific behavior of an organism that recurs with a regularity of approximately 24 hours. |
| circadian regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression such that an expression pattern recurs with a regularity of approximately 24 hours. |
| circadian rhythm | Any biological process in an organism that recurs with a regularity of approximately 24 hours. |
| endocytosis | A vesicle-mediated transport process in which cells take up external materials or membrane constituents by the invagination of a small region of the plasma membrane to form a new membrane-bounded vesicle. |
| negative regulation of Wnt signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of the Wnt signaling pathway. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of amyloid-beta formation | Any process that activates or increases the frequency, rate or extent of amyloid-beta formation. |
| positive regulation of canonical Wnt signaling pathway | Any process that increases the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| positive regulation of non-canonical Wnt signaling pathway | Any process that activates or increases the frequency, rate or extent of non-canonical Wnt-activated signaling pathway. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| positive regulation of Wnt signaling pathway | Any process that activates or increases the frequency, rate or extent of Wnt signal transduction. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of circadian rhythm | Any process that modulates the frequency, rate or extent of a circadian rhythm. A circadian rhythm is a biological process in an organism that recurs with a regularity of approximately 24 hours. |
| regulation of protein localization | Any process that modulates the frequency, rate or extent of any process in which a protein is transported to, or maintained in, a specific location. |
| Wnt signaling pathway | The series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell and ending with a change in cell state. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P29295 | HRR25 | Casein kinase I homolog HRR25 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P35508 | CSNK1D | Casein kinase I isoform delta | Bos taurus (Bovine) | SS |
| Q5ZLL1 | CSNK1E | Casein kinase I isoform epsilon | Gallus gallus (Chicken) | SS |
| Q86Y07 | VRK2 | Serine/threonine-protein kinase VRK2 | Homo sapiens (Human) | PR |
| P48730 | CSNK1D | Casein kinase I isoform delta | Homo sapiens (Human) | SS |
| P49674 | CSNK1E | Casein kinase I isoform epsilon | Homo sapiens (Human) | EV |
| Q9DC28 | Csnk1d | Casein kinase I isoform delta | Mus musculus (Mouse) | PR |
| Q8BN21 | Vrk2 | Serine/threonine-protein kinase VRK2 | Mus musculus (Mouse) | PR |
| Q06486 | Csnk1d | Casein kinase I isoform delta | Rattus norvegicus (Rat) | PR |
| P42169 | C03C10.2 | Putative casein kinase I C03C10.2 | Caenorhabditis elegans | PR |
| P34516 | K06H7.8 | Putative serine/threonine-protein kinase K06H7.1 | Caenorhabditis elegans | PR |
| Q6P647 | csnk1d | Casein kinase I isoform delta | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q7T2E3 | csnk1da | Casein kinase I isoform delta-A | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q7ZUS1 | vrk1 | Serine/threonine-protein kinase VRK1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q6P3K7 | csnk1db | Casein kinase I isoform delta-B | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MELRVGNKYR | LGRKIGSGSF | GDIYLGANIA | SGEEVAIKLE | CVKTKHPQLH | IESKFYKMMQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GGVGIPSIKW | CGAEGDYNVM | VMELLGPSLE | DLFNFCSRKF | SLKTVLLLAD | QMISRIEYIH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SKNFIHRDVK | PDNFLMGLGK | KGNLVYIIDF | GLAKKYRDAR | THQHIPYREN | KNLTGTARYA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SINTHLGIEQ | SRRDDLESLG | YVLMYFNLGS | LPWQGLKAAT | KRQKYERISE | KKMSTPIEVL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| CKGYPSEFST | YLNFCRSLRF | DDKPDYSYLR | QLFRNLFHRQ | GFSYDYVFDW | NMLKFGAARN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PEDVDRERRE | HEREERMGQL | RGSATRALPP | GPPTGATANR | LRSAAEPVAS | TPASRIQQTG |
| 370 | 380 | 390 | 400 | 410 | |
| NTSPRAISRA | DRERKVSMRL | HRGAPANVSS | SDLTGRQEVS | RLAASQTSVP | FDHLGK |