Q9JKF1
Gene name |
Iqgap1 |
Protein name |
Ras GTPase-activating-like protein IQGAP1 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:29875 |
EC number |
|
Protein Class |
|
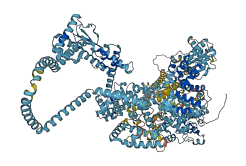
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
956-1274 (C1 fragments) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for Q9JKF1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9JKF1-F1 | Predicted | AlphaFoldDB |
7 variants for Q9JKF1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs232712612 | 92 | L>V | No | Ensembl | |
| rs216296629 | 322 | V>G | No | Ensembl | |
| rs33220621 | 373 | A>V | No | Ensembl | |
| rs13471640 | 1006 | I>M | No | Ensembl | |
| rs233736618 | 1268 | V>A | No | Ensembl | |
| rs13471636 | 1268 | V>L | No | Ensembl | |
| rs244359156 | 1551 | M>I | No | Ensembl |
No associated diseases with Q9JKF1
9 regional properties for Q9JKF1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| binding_site | IQ motif, EF-hand binding site | 744 - 774 | IPR000048-1 |
| binding_site | IQ motif, EF-hand binding site | 774 - 804 | IPR000048-2 |
| binding_site | IQ motif, EF-hand binding site | 804 - 834 | IPR000048-3 |
| binding_site | IQ motif, EF-hand binding site | 834 - 864 | IPR000048-4 |
| domain | RasGAP protein, C-terminal | 1452 - 1580 | IPR000593 |
| domain | WW domain | 685 - 712 | IPR001202 |
| domain | Calponin homology domain | 44 - 159 | IPR001715 |
| domain | Ras GTPase-activating domain | 992 - 1345 | IPR001936 |
| conserved_site | Ras GTPase-activating protein, conserved site | 1189 - 1203 | IPR023152 |
Functions
20 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| cell leading edge | The area of a motile cell closest to the direction of movement. |
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| cortical actin cytoskeleton | The portion of the actin cytoskeleton, comprising filamentous actin and associated proteins, that lies just beneath the plasma membrane. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic ribonucleoprotein granule | A ribonucleoprotein granule located in the cytoplasm. |
| extrinsic component of cytoplasmic side of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to its cytoplasmic surface, but not integrated into the hydrophobic region. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| growth cone | The migrating motile tip of a growing neuron projection, where actin accumulates, and the actin cytoskeleton is the most dynamic. |
| lateral plasma membrane | The portion of the plasma membrane at the lateral side of the cell. In epithelial cells, lateral plasma membranes are on the sides of cells which lie at the interface of adjacent cells. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| ribonucleoprotein complex | A macromolecular complex that contains both RNA and protein molecules. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
| slit diaphragm | A specialized cell-cell junction found between the interdigitating foot processes of the glomerular epithelium (the podocytes) in the vertebrate kidney, which is adapted for facilitating glomerular filtration. |
15 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| GTPase activator activity | Binds to and increases the activity of a GTPase, an enzyme that catalyzes the hydrolysis of GTP. |
| MAP-kinase scaffold activity | The binding activity of a molecule that functions as a physical support for the assembly of a multiprotein mitogen-activated protein kinase (MAPK) complex. Binds multiple kinases of the MAPKKK cascade, and also upstream signaling proteins, permitting those molecules to function in a coordinated way. Bringing together multiple enzymes and their substrates enables the signal to be transduced quickly and efficiently. |
| mitogen-activated protein kinase binding | Binding to a mitogen-activated protein kinase. |
| molecular adaptor activity | The binding activity of a molecule that brings together two or more molecules through a selective, non-covalent, often stoichiometric interaction, permitting those molecules to function in a coordinated way. |
| phosphatidylinositol-3,4,5-trisphosphate binding | Binding to phosphatidylinositol-3,4,5-trisphosphate, a derivative of phosphatidylinositol in which the inositol ring is phosphorylated at the 3', 4' and 5' positions. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein phosphatase binding | Binding to a protein phosphatase. |
| protein serine/threonine kinase activator activity | Binds to and increases the activity of a protein serine/threonine kinase. |
| protein-containing complex binding | Binding to a macromolecular complex. |
| S100 protein binding | Binding to a S100 protein. S100 is a small calcium and zinc binding protein produced in astrocytes that is implicated in Alzheimer's disease, Down Syndrome and ALS. |
| small GTPase binding | Binding to a small monomeric GTPase. |
25 GO annotations of biological process
| Name | Definition |
|---|---|
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cellular response to calcium ion | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a calcium ion stimulus. |
| cellular response to epidermal growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an epidermal growth factor stimulus. |
| cellular response to fibroblast growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an fibroblast growth factor stimulus. |
| cellular response to platelet-derived growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a platelet-derived growth factor stimulus. |
| epidermal growth factor receptor signaling pathway | The series of molecular signals initiated by binding of a ligand to the tyrosine kinase receptor EGFR (ERBB1) on the surface of a cell. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| fibroblast growth factor receptor signaling pathway | The series of molecular signals generated as a consequence of a fibroblast growth factor receptor binding to one of its physiological ligands. |
| fibroblast migration | Cell migration that is accomplished by extension and retraction of a fibroblast pseudopodium. A fibroblast is a connective tissue cell which secretes an extracellular matrix rich in collagen and other macromolecules. |
| glomerular visceral epithelial cell development | The process whose specific outcome is the progression of a glomerular visceral epithelial cell over time, from its formation to the mature structure. A glomerular visceral epithelial cell is a specialized epithelial cell that contains 'feet' that interdigitate with the 'feet' of other glomerular epithelial cells. |
| negative regulation of dephosphorylation | Any process the stops, prevents, or reduces the frequency, rate or extent of removal of phosphate groups from a molecule. |
| neuron projection extension | Long distance growth of a single neuron projection involved in cellular development. A neuron projection is a prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| platelet-derived growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a platelet-derived growth factor receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| positive regulation of dendrite development | Any process that activates or increases the frequency, rate or extent of dendrite development. |
| positive regulation of focal adhesion assembly | Any process that activates or increases the frequency, rate or extent of focal adhesion assembly, the establishment and maturation of focal adhesions. |
| positive regulation of MAP kinase activity | Any process that activates or increases the frequency, rate or extent of MAP kinase activity. |
| positive regulation of MAPK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade. |
| positive regulation of peptidyl-tyrosine autophosphorylation | Any process that activates or increases the frequency, rate or extent of peptidyl-tyrosine autophosphorylation. |
| positive regulation of protein kinase activity | Any process that activates or increases the frequency, rate or extent of protein kinase activity. |
| positive regulation of protein localization | Any process that activates or increases the frequency, rate or extent of a protein localization. |
| positive regulation of vascular associated smooth muscle cell migration | Any process that activates or increases the frequency, rate or extent of vascular associated smooth muscle cell migration. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of cytokine production | Any process that modulates the frequency, rate, or extent of production of a cytokine. |
| regulation of GTPase activity | Any process that modulates the rate of GTP hydrolysis by a GTPase. |
| regulation of mitotic cell cycle | Any process that modulates the rate or extent of progress through the mitotic cell cycle. |
| response to angiotensin | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an angiotensin stimulus. Angiotensin is any of three physiologically active peptides (angiotensin II, III, or IV) processed from angiotensinogen. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q13576 | IQGAP2 | Ras GTPase-activating-like protein IQGAP2 | Homo sapiens (Human) | SS |
| P46940 | IQGAP1 | Ras GTPase-activating-like protein IQGAP1 | Homo sapiens (Human) | EV |
| Q86VI3 | IQGAP3 | Ras GTPase-activating-like protein IQGAP3 | Homo sapiens (Human) | SS |
| Q3UQ44 | Iqgap2 | Ras GTPase-activating-like protein IQGAP2 | Mus musculus (Mouse) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSAAEEVDGL | GVVRPHYGSV | LDNERLTAEE | MDERRRQNVA | YEYLCHLEEA | KRWMEACLGE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DLPPTTELEE | GLRNGVYLAK | LGNFFSPKVV | SLKKIYDREQ | TRYKATGLHF | RHTDNVIQWL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NAMDEIGLPK | IFYPETTDIY | DRKNMPRCIY | CIHALSLYLF | KLGLAPQIQD | LYGKVDFTEE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EINNMKIELE | KYGIQMPAFS | KIGGILANEL | SVDEAALHAA | VIAINEAIDR | RVAADTFTAL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KNPNAMLVNL | EEGLAPTYQD | VLYQAKQDKM | TNAKNRTENS | DRERDVYEEL | LTQAEIQGNV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NKVNTSSALA | NISLALEQGC | AVTLLKALQS | LALGLRGLQT | QNSDWYMKQL | QSDLQQKRQS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GQTDPLQKEE | VQAGVDAANS | AAQQYQRRLA | AVAAINAAIQ | KGIAEKTVLE | LMNPEAQLPQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VYPFAADLYQ | KELATLQQQS | PEHSLTHPEL | TVAVEMLSSV | ALINRALESG | DMTTVWKQLS |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SSVTGLTNIE | EENCQRYLDE | LMKLKAQAHA | ENNAFITWND | IQACVDHVNL | VVHEEHERIL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| AIGLINEALD | EGDAQKTLQA | LQIPAAKLEG | VLAEVAQHYQ | DTLIRAKREK | AQETQDESAV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LWLDEIQGGI | WQSNKDTQEA | QRFALGISAI | NEAVDSGDVG | RTLSALRSPD | VGLYGVIPEC |
| 670 | 680 | 690 | 700 | 710 | 720 |
| GETYQSDLAE | AKKKRLAAGD | NNSKWVKHWV | KGGYHYYHNL | ETQAGGWAEP | PDFVQNSVQL |
| 730 | 740 | 750 | 760 | 770 | 780 |
| SREEIQSSIS | GVTAAYNREQ | LWLANEGLIT | KLQACCRGYL | VRQEFRSRMN | FLKKQIPAIT |
| 790 | 800 | 810 | 820 | 830 | 840 |
| CIQSQWRGYK | QKKAYQDRLA | YLHSHKDEVV | KIQSLARMHQ | ARKRYRDRLQ | YFRDHINDII |
| 850 | 860 | 870 | 880 | 890 | 900 |
| KIQAFIRANK | ARDDYKTLIN | AEDPPMIVVR | KFVHLLDQSD | QDFQEELDLM | KMREEVITLI |
| 910 | 920 | 930 | 940 | 950 | 960 |
| RSNQQLENDL | NLMDIKIGLL | VKNKITLQDV | VSHSKKLTKK | NKEQLSDMMM | INKQKGGLKA |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| LSKEKREKLE | AYQHLFYLLQ | TNPTYLAKLI | FQMPQNKSTK | FMDSVIFTLY | NYASNQREEY |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| LLLRLFQTAL | QEEIKSKVDQ | IQEIVTGNPT | VIKMVVSFNR | GARGQNALRQ | ILAPVVKEIM |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| DDKSLNIKTD | PVDIYKSWVN | QMESQTGEAS | KLPYDVTPEQ | ALSHEEVKTR | LDNSIRNMRA |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| VTDKFLSAIV | SSVDKIPYGM | RFIAKVLKDS | LHEKFPDAGE | DELLKIIGNL | LYYRYMNPAI |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| VAPDAFDIID | LSAGGQLTTD | QRRNLGSIAK | MLQHAASNKM | FLGDNAHLSI | INEYLSQSYQ |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| KFRRFFQVAC | DVPELQDKFN | VDEYSDLVTL | TKPVIYISIG | EIINTHTLLL | DHQDAIAPEH |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| NDPIHELLDD | LGEVPTIESL | IGESCGNSND | PNKEALAKTE | VSLTLTNKFD | VPGDENAEMD |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| ARTILLNTKR | LIVDVIRFQP | GETLTEILET | PATNEQEAEH | QRAMQRRAIR | DAKTPDKMKK |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| SKPMKEDNNL | SLQEKKEKIQ | TGLKKLTELG | TVDPKNRYQE | LINDIAKDIR | NQRRYRQRRK |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| AELVKLQQTY | SALNSKATFY | GEQVDYYKSY | IKTCLDNLAS | KGKVSKKPRE | MKGKKSKKIS |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| LKYTAARLHE | KGVLLEIEDL | QANQFKNVIF | EIGPTEEVGD | FEVKAKFMGV | QMETFMLHYQ |
| 1630 | 1640 | 1650 | |||
| DLLQLQYEGV | AVMKLFDRAK | VNVNLLIFLL | NKKFYGK |