Q9JK66
Gene name |
Prkn |
Protein name |
E3 ubiquitin-protein ligase parkin |
Names |
Parkin RBR E3 ubiquitin-protein ligase |
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
rno:56816 |
EC number |
2.3.2.31: Aminoacyltransferases |
Protein Class |
RBR FAMILY RING FINGER AND IBR DOMAIN-CONTAINING (PTHR11685) |
Descriptions
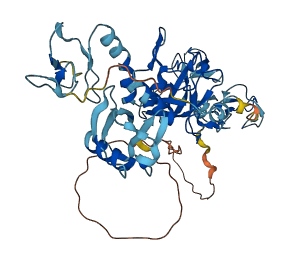
Parkin belongs to the RBR subfamily of E3 ubiquitin ligases that function by transferring Ub from an E2-conjugating enzyme to a substrate through an E3-Ub intermediate. Parkin contains an N-terminal ubiquitin-like (UbL) domain connected through a linker to four zinc-coordinating domains consisting of RING0, following by the RBR (RING1, In-Between-RING (IBR), RING2) module common to other E3 Ub ligases. RING1 contains an E2 binding site and RING2 contains a catalytic cysteine that is an acceptor for an ubiquitin from RING1-bound E2~Ub forming an intermediate (E3~Ub) that leads to substrate or autoubiquitination.
Structural studies revealed that Parkin, in its basal state, adopts an autoinhibited conformation with several distinct sites of inhibition. First, the UbL domain is bound to RING1 such that Ser65 is poorly accessible for phosphorylation by PINK1. Second, the Repressor Element of Parkin (REP) blocks the site of E2 Ub-conjugating enzyme binding on RING1. Finally, the RING0 domain binds to RING2, where it occludes the Cys431 Ub acceptor site. PINK1 stimulates parkin activity through phosphorylation of both Ub and parkin's UbL domain at an equivalent serine 65 position in sequence and structure. The phosphorylation of both the UbL domain and Ub are required to activate parkin by releasing the UbL domain, forming an extended structure needed to facilitate E2-Ub binding.
Autoinhibitory domains (AIDs)
Target domain |
228-318 (RING1 domain) |
Relief mechanism |
PTM |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Target domain |
228-318 (RING1 domain); 409-463 (RING2 domain) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
409-463 (RING2 domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
- Byrd RA et al. (2013) "Compact Parkin only: insights into the structure of an autoinhibited ubiquitin ligase", The EMBO journal, 32, 2087-9
- Aguirre JD et al. (2017) "Structure of phosphorylated UBL domain and insights into PINK1-orchestrated parkin activation", Proceedings of the National Academy of Sciences of the United States of America, 114, 298-303
- Kumar A et al. (2015) "Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis", The EMBO journal, 34, 2506-21
- Tang MY et al. (2017) "Structure-guided mutagenesis reveals a hierarchical mechanism of Parkin activation", Nature communications, 8, 14697
- Seirafi M et al. (2015) "Parkin structure and function", The FEBS journal, 282, 2076-88
Autoinhibited structure

Activated structure

1 variants for Q9JK66
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs107030066 | 138 | E>A | No | Ensembl |
No associated diseases with Q9JK66
6 regional properties for Q9JK66
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | CARD domain | 453 - 543 | IPR001315 |
| repeat | BIR repeat | 44 - 115 | IPR001370-1 |
| repeat | BIR repeat | 182 - 252 | IPR001370-2 |
| repeat | BIR repeat | 267 - 338 | IPR001370-3 |
| domain | Zinc finger, RING-type | 571 - 606 | IPR001841 |
| domain | BIRC2/BIRC3, UBA domain | 391 - 437 | IPR041933 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.31 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11685 | RBR FAMILY RING FINGER AND IBR DOMAIN-CONTAINING |
| PANTHER Subfamily | PTHR11685:SF471 | E3 UBIQUITIN-PROTEIN LIGASE PARKIN |
| PANTHER Protein Class |
ubiquitin-protein ligase
protein modifying enzyme |
|
| PANTHER Pathway Category |
Parkinson disease Parkin |
|
26 GO annotations of cellular component
| Name | Definition |
|---|---|
| aggresome | An inclusion body formed by dynein-dependent retrograde transport of an aggregated protein on microtubules. |
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| intracellular vesicle | Any vesicle that is part of the intracellular region. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| mitochondrial outer membrane | The outer, i.e. cytoplasm-facing, lipid bilayer of the mitochondrial envelope. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| Parkin-FBXW7-Cul1 ubiquitin ligase complex | A ubiquitin ligase complex containing Parkin (PARK2), the F-box protein FBXW7 (also called SEL-10) and a cullin from the Cul1 subfamily; substrate specificity is conferred by the F-box protein. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
| synaptic vesicle membrane | The lipid bilayer surrounding a synaptic vesicle. |
| terminal bouton | Terminal inflated portion of the axon, containing the specialized apparatus necessary to release neurotransmitters. The axon terminus is considered to be the whole region of thickening and the terminal bouton is a specialized region of it. |
| ubiquitin ligase complex | A protein complex that includes a ubiquitin-protein ligase and enables ubiquitin protein ligase activity. The complex also contains other proteins that may confer substrate specificity on the complex. |
25 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| beta-catenin binding | Binding to a catenin beta subunit. |
| chaperone binding | Binding to a chaperone protein, a class of proteins that bind to nascent or unfolded polypeptides and ensure correct folding or transport. |
| cullin family protein binding | Binding to a member of the cullin family, hydrophobic proteins that act as scaffolds for ubiquitin ligases (E3). |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| F-box domain binding | Binding to an F-box domain of a protein. |
| G protein-coupled receptor binding | Binding to a G protein-coupled receptor. |
| heat shock protein binding | Binding to a heat shock protein, a protein synthesized or activated in response to heat shock. |
| histone deacetylase binding | Binding to histone deacetylase. |
| Hsp70 protein binding | Binding to a Hsp70 protein, heat shock proteins around 70kDa in size. |
| identical protein binding | Binding to an identical protein or proteins. |
| kinase binding | Binding to a kinase, any enzyme that catalyzes the transfer of a phosphate group. |
| metal ion binding | Binding to a metal ion. |
| PDZ domain binding | Binding to a PDZ domain of a protein, a domain found in diverse signaling proteins. |
| phospholipase binding | Binding to a phospholipase. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein-containing complex binding | Binding to a macromolecular complex. |
| transcription corepressor activity | A transcription coregulator activity that represses or decreases the transcription of specific gene sets via binding to a DNA-bound DNA-binding transcription factor, either on its own or as part of a complex. Corepressors often act by altering chromatin structure and modifications. For example, one class of transcription corepressors modifies chromatin structure through covalent modification of histones. A second class remodels the conformation of chromatin in an ATP-dependent fashion. A third class modulates interactions of DNA-bound DNA-binding transcription factors with other transcription coregulators. |
| tubulin binding | Binding to monomeric or multimeric forms of tubulin, including microtubules. |
| ubiquitin binding | Binding to ubiquitin, a protein that when covalently bound to other cellular proteins marks them for proteolytic degradation. |
| ubiquitin conjugating enzyme binding | Binding to a ubiquitin conjugating enzyme, any of the E2 proteins. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
| ubiquitin-specific protease binding | Binding to a ubiquitin-specific protease. |
110 GO annotations of biological process
| Name | Definition |
|---|---|
| adult locomotory behavior | Locomotory behavior in a fully developed and mature organism. |
| aggresome assembly | The aggregation, arrangement and bonding together of a set of components to form an aggresome; requires the microtubule cytoskeleton and dynein. |
| autophagy of mitochondrion | The autophagic process in which mitochondria are delivered to a type of vacuole and degraded in response to changing cellular conditions. |
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| cellular response to hydrogen sulfide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hydrogen sulfide stimulus. |
| cellular response to L-glutamate | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a L-glutamate(1-) stimulus. |
| cellular response to L-glutamine | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a L-glutamine stimulus. |
| cellular response to manganese ion | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a manganese ion stimulus. |
| cellular response to toxic substance | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a toxic stimulus. |
| dopamine metabolic process | The chemical reactions and pathways involving dopamine, a catecholamine neurotransmitter and a metabolic precursor of noradrenaline and adrenaline. |
| dopamine uptake involved in synaptic transmission | The directed movement of dopamine into a presynaptic neuron or glial cell. In this context, dopamine is a catecholamine neurotransmitter and a metabolic precursor of noradrenaline and adrenaline. |
| free ubiquitin chain polymerization | The process of creating free ubiquitin chains, compounds composed of a large number of ubiquitin monomers. These chains are not conjugated to a protein. |
| learning | Any process in an organism in which a relatively long-lasting adaptive behavioral change occurs as the result of experience. |
| locomotory behavior | The specific movement from place to place of an organism in response to external or internal stimuli. Locomotion of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions. |
| mitochondrial fission | The division of a mitochondrion within a cell to form two or more separate mitochondrial compartments. |
| mitochondrial fragmentation involved in apoptotic process | The change in the morphology of the mitochondria in an apoptotic cell from a highly branched network to a fragmented vesicular form. |
| mitochondrion localization | Any process in which a mitochondrion or mitochondria are transported to, and/or maintained in, a specific location within the cell. |
| mitochondrion organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a mitochondrion; includes mitochondrial morphogenesis and distribution, and replication of the mitochondrial genome as well as synthesis of new mitochondrial components. |
| mitochondrion to lysosome transport | Transport from the mitochondrion to the lysosome, mediated by mitochondrion-derived vesicles. |
| mitophagy | The selective autophagy process in which a mitochondrion is degraded by macroautophagy. |
| modulation of chemical synaptic transmission | Any process that modulates the frequency or amplitude of synaptic transmission, the process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse. Amplitude, in this case, refers to the change in postsynaptic membrane potential due to a single instance of synaptic transmission. |
| negative regulation by host of viral genome replication | A process in which a host organism stops, prevents or reduces the frequency, rate or extent of viral genome replication. |
| negative regulation of actin filament bundle assembly | Any process that stops, prevents, or reduces the frequency, rate or extent of the assembly of actin filament bundles. |
| negative regulation of canonical Wnt signaling pathway | Any process that decreases the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of an endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway. |
| negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of an endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway. |
| negative regulation of excitatory postsynaptic potential | Any process that prevents the establishment or decreases the extent of the excitatory postsynaptic potential (EPSP) which is a temporary increase in postsynaptic potential due to the flow of positively charged ions into the postsynaptic cell. The flow of ions that causes an EPSP is an excitatory postsynaptic current (EPSC) and makes it easier for the neuron to fire an action potential. |
| negative regulation of exosomal secretion | Any process that stops, prevents or reduces the frequency, rate or extent of exosomal secretion. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| negative regulation of glucokinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of glucokinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a glucose molecule. |
| negative regulation of insulin secretion | Any process that stops, prevents, or reduces the frequency, rate or extent of the regulated release of insulin. |
| negative regulation of intralumenal vesicle formation | Any process that stops, prevents or reduces the frequency, rate or extent of intralumenal vesicle formation. |
| negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | Any process that stops, prevents or reduces the frequency, rate or extent of intrinsic apoptotic signaling pathway by p53 class mediator. |
| negative regulation of JNK cascade | Any process that stops, prevents, or reduces the frequency, rate or extent of signal transduction mediated by the JNK cascade. |
| negative regulation of mitochondrial fission | Any process that decreases the rate, frequency or extent of mitochondrial fission. Mitochondrial fission is the division of a mitochondrion within a cell to form two or more separate mitochondrial compartments. |
| negative regulation of mitochondrial fusion | Any process that decreases the frequency, rate or extent of merging of two or more mitochondria within a cell to form a single compartment. |
| negative regulation of neuron apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process in neurons. |
| negative regulation of neuron death | Any process that stops, prevents or reduces the frequency, rate or extent of neuron death. |
| negative regulation of oxidative stress-induced neuron death | Any process that stops, prevents or reduces the frequency, rate or extent of oxidative stress-induced neuron death. |
| negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of oxidative stress-induced neuron intrinsic apoptotic signaling pathway. |
| negative regulation of primary amine oxidase activity | Any process that stops, prevents or reduces the frequency, rate or extent of primary amine oxidase activity. |
| negative regulation of protein phosphorylation | Any process that stops, prevents or reduces the rate of addition of phosphate groups to amino acids within a protein. |
| negative regulation of reactive oxygen species biosynthetic process | Any process that stops, prevents or reduces the frequency, rate or extent of reactive oxygen species biosynthetic process. |
| negative regulation of reactive oxygen species metabolic process | Any process that stops, prevents or reduces the frequency, rate or extent of reactive oxygen species metabolic process. |
| negative regulation of release of cytochrome c from mitochondria | Any process that decreases the rate, frequency or extent of release of cytochrome c from mitochondria, the process in which cytochrome c is enabled to move from the mitochondrial intermembrane space into the cytosol, which is an early step in apoptosis and leads to caspase activation. |
| negative regulation of spontaneous neurotransmitter secretion | Any process that stops, prevents or reduces the frequency, rate or extent of spontaneous neurotransmitter secretion. |
| negative regulation of synaptic transmission, glutamatergic | Any process that stops, prevents, or reduces the frequency, rate or extent of glutamatergic synaptic transmission, the process of communication from a neuron to another neuron across a synapse using the neurotransmitter glutamate. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| neuron cellular homeostasis | The cellular homeostatic process that preserves a neuron in a stable, differentiated functional and structural state. |
| norepinephrine metabolic process | The chemical reactions and pathways involving norepinephrine, a hormone secreted by the adrenal medulla, and a neurotransmitter in the sympathetic peripheral nervous system and in some tracts in the central nervous system. It is also the demethylated biosynthetic precursor of epinephrine. |
| parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | A positive regulation of the macromitophagy pathway that is triggered by mitochondrial depolarization and requires the function of a parkin-family molecule. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of ATP biosynthetic process | Any process that activates or increases the frequency, rate or extent of ATP biosynthetic process. |
| positive regulation of autophagy of mitochondrion | Any process that activates or increases the frequency, rate or extent of mitochondrion degradation by autophagy. |
| positive regulation of dendrite extension | Any process that activates or increases the frequency, rate or extent of dendrite extension. |
| positive regulation of DNA binding | Any process that increases the frequency, rate or extent of DNA binding. DNA binding is any process in which a gene product interacts selectively with DNA (deoxyribonucleic acid). |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of I-kappaB kinase/NF-kappaB signaling | Any process that activates or increases the frequency, rate or extent of I-kappaB kinase/NF-kappaB signaling. |
| positive regulation of insulin secretion involved in cellular response to glucose stimulus | Any process that increases the frequency, rate or extent of the regulated release of insulin that contributes to the response of a cell to glucose. |
| positive regulation of mitochondrial fission | Any process that increases the rate, frequency or extent of mitochondrial fission. Mitochondrial fission is the division of a mitochondrion within a cell to form two or more separate mitochondrial compartments. |
| positive regulation of mitochondrial fusion | Any process that increases the frequency, rate or extent of merging of two or more mitochondria within a cell to form a single compartment. |
| positive regulation of mitochondrial membrane potential | Any process that activates or increases the frequency, rate or extent of establishment or extent of a mitochondrial membrane potential, the electric potential existing across any mitochondrial membrane arising from charges in the membrane itself and from the charges present in the media on either side of the membrane. |
| positive regulation of mitophagy | Any process that activates or increases the frequency, rate or extent of mitophagy. |
| positive regulation of mitophagy in response to mitochondrial depolarization | Any process that activates or increases the frequency, rate or extent of mitophagy in response to mitochondrial depolarization. |
| positive regulation of neurotransmitter uptake | Any process that activates or increases the frequency, rate or extent of the directed movement of a neurotransmitter into a neuron or glial cell. |
| positive regulation of proteasomal protein catabolic process | Any process that activates or increases the frequency, rate or extent of proteasomal protein catabolic process. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| positive regulation of protein binding | Any process that activates or increases the frequency, rate or extent of protein binding. |
| positive regulation of protein linear polyubiquitination | Any process that activates or increases the frequency, rate or extent of protein linear polyubiquitination. |
| positive regulation of protein localization to membrane | Any process that activates or increases the frequency, rate or extent of protein localization to membrane. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of tumor necrosis factor-mediated signaling pathway | Any process that activates or increases the frequency, rate or extent of tumor necrosis factor-mediated signaling pathway. |
| proteasomal protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds that is mediated by the proteasome. |
| proteasome-mediated ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| protein autoubiquitination | The ubiquitination by a protein of one or more of its own amino acid residues, or residues on an identical protein. Ubiquitination occurs on the lysine residue by formation of an isopeptide crosslink. |
| protein destabilization | Any process that decreases the stability of a protein, making it more vulnerable to degradative processes or aggregation. |
| protein K11-linked ubiquitination | A protein ubiquitination process in which ubiquitin monomers are attached to a protein, and then ubiquitin polymers are formed by linkages between lysine residues at position 11 of the ubiquitin monomers. K11-linked polyubiquitination targets the substrate protein for degradation. The anaphase-promoting complex promotes the degradation of mitotic regulators by assembling K11-linked polyubiquitin chains. |
| protein K48-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 48 of the ubiquitin monomers, is added to a protein. K48-linked ubiquitination targets the substrate protein for degradation. |
| protein K6-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 6 of the ubiquitin monomers, is added to a protein. K6-linked ubiquitination is involved in DNA repair. |
| protein K63-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 63 of the ubiquitin monomers, is added to a protein. K63-linked ubiquitination does not target the substrate protein for degradation, but is involved in several pathways, notably as a signal to promote error-free DNA postreplication repair. |
| protein localization to mitochondrion | A process in which a protein is transported to, or maintained in, a location within the mitochondrion. |
| protein metabolic process | The chemical reactions and pathways involving a protein. Includes protein modification. |
| protein monoubiquitination | Addition of a single ubiquitin group to a protein. |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| protein stabilization | Any process involved in maintaining the structure and integrity of a protein and preventing it from degradation or aggregation. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| regulation of apoptotic process | Any process that modulates the occurrence or rate of cell death by apoptotic process. |
| regulation of autophagy | Any process that modulates the frequency, rate or extent of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| regulation of cellular response to oxidative stress | Any process that modulates the frequency, rate or extent of cellular response to oxidative stress. |
| regulation of dopamine metabolic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving dopamine. |
| regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| regulation of mitochondrial membrane potential | Any process that modulates the establishment or extent of the mitochondrial membrane potential, the electric potential existing across the mitochondrial membrane arising from charges in the membrane itself and from the charges present in the media on either side of the membrane. |
| regulation of mitochondrion organization | Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of a mitochondrion. |
| regulation of neurotransmitter secretion | Any process that modulates the frequency, rate or extent of the regulated release of a neurotransmitter from a cell. |
| regulation of protein stability | Any process that affects the structure and integrity of a protein, altering the likelihood of its degradation or aggregation. |
| regulation of protein ubiquitination | Any process that modulates the frequency, rate or extent of the addition of ubiquitin groups to a protein. |
| regulation of reactive oxygen species metabolic process | Any process that modulates the frequency, rate or extent of reactive oxygen species metabolic process. |
| response to corticosterone | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a corticosterone stimulus. Corticosterone is a 21 carbon steroid hormone of the corticosteroid type, produced in the cortex of the adrenal glands. In many species, corticosterone is the principal glucocorticoid, involved in regulation of fuel metabolism, immune reactions, and stress responses. |
| response to curcumin | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a curcumin stimulus. |
| response to endoplasmic reticulum stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stress acting at the endoplasmic reticulum. ER stress usually results from the accumulation of unfolded or misfolded proteins in the ER lumen. |
| response to muscle activity | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a muscle activity stimulus. |
| response to oxidative stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| response to unfolded protein | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an unfolded protein stimulus. |
| response to xenobiotic stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a xenobiotic, a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
| startle response | An action or movement due to the application of a sudden unexpected stimulus. |
| synaptic transmission, dopaminergic | The vesicular release of dopamine. from a presynapse, across a chemical synapse, the subsequent activation of dopamine receptors at the postsynapse of a target cell (neuron, muscle, or secretory cell) and the effects of this activation on the postsynaptic membrane potential and ionic composition of the postsynaptic cytosol. This process encompasses both spontaneous and evoked release of neurotransmitter and all parts of synaptic vesicle exocytosis. Evoked transmission starts with the arrival of an action potential at the presynapse. |
| synaptic transmission, glutamatergic | The vesicular release of glutamate from a presynapse, across a chemical synapse, the subsequent activation of glutamate receptors at the postsynapse of a target cell (neuron, muscle, or secretory cell) and the effects of this activation on the postsynaptic membrane potential and ionic composition of the postsynaptic cytosol. This process encompasses both spontaneous and evoked release of neurotransmitter and all parts of synaptic vesicle exocytosis. Evoked transmission starts with the arrival of an action potential at the presynapse. |
| ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of a ubiquitin group, or multiple ubiquitin groups, to the protein. |
| zinc ion homeostasis | Any process involved in the maintenance of an internal steady state of zinc ions within an organism or cell. |
3 homologous proteins in AiPD
| 10 | 20 | 30 | 40 | 50 | 60 |
| MIVFVRFNSS | YGFPVEVDSD | TSIFQLKEVV | AKRQGVPADQ | LRVIFAGKEL | QNHLTVQNCD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LEQQSIVHIV | QRPQRKSHET | NASGGDKPQS | TPEGSIWEPR | SLTRVDLSSH | ILPADSVGLA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VILDTDSKSD | SEAARGPEAK | PTYHSFFVYC | KGPCHKVQPG | KLRVQCGTCR | QATLTLAQGP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SCWDDVLIPN | RMSGECQSPD | CPGTRAEFFF | KCGAHPTSDK | DTSVALNLIT | NNSRSIPCIA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| CTDVRNPVLV | FQCNHRHVIC | LDCFHLYCVT | RLNDRQFVHD | AQLGYSLPCV | AGCPNSLIKE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LHHFRILGEE | QYNRYQQYGA | EECVLQMGGV | LCPRPGCGAG | LLPEQGQKKV | TCEGGNGLGC |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GFVFCRDCKE | AYHEGECDSM | FEASGATSQA | YRVDQRAAEQ | ARWEEASKET | IKKTTKPCPR |
| 430 | 440 | 450 | 460 | ||
| CNVPIEKNGG | CMHMKCPQPQ | CKLEWCWNCG | CEWNRACMGD | HWFDV |