Q9JK48
Gene name |
Sh3glb1 (Kiaa0491) |
Protein name |
Endophilin-B1 |
Names |
SH3 domain-containing GRB2-like protein B1 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:54673 |
EC number |
|
Protein Class |
|
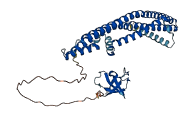
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q9JK48
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1X43 | NMR | - | A | 298-365 | PDB |
| AF-Q9JK48-F1 | Predicted | AlphaFoldDB |
22 variants for Q9JK48
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388665451 | 50 | E>* | No | EVA | |
| rs3388663321 | 70 | N>K | No | EVA | |
| rs3388664495 | 94 | P>S | No | EVA | |
| rs3388664500 | 96 | L>V | No | EVA | |
| rs3388655432 | 169 | R>* | No | EVA | |
| rs3388665434 | 170 | L>V | No | EVA | |
| rs3388649343 | 188 | K>* | No | EVA | |
| rs3388664480 | 192 | E>K | No | EVA | |
| rs3388655460 | 202 | F>V | No | EVA | |
| rs3400428550 | 230 | F>L | No | EVA | |
| rs3393817819 | 268 | G>W | No | EVA | |
| rs3393817802 | 270 | P>L | No | EVA | |
| rs3388665503 | 270 | P>S | No | EVA | |
| rs3388667997 | 280 | G>C | No | EVA | |
| rs3388670492 | 283 | A>P | No | EVA | |
| rs214480570 | 287 | T>A | No | EVA | |
| rs3393818928 | 297 | N>H | No | EVA | |
| rs31151155 | 307 | N>S | No | EVA | |
| rs3388667990 | 344 | W>C | No | EVA | |
| rs3388657973 | 345 | L>V | No | EVA | |
| rs3388670522 | 357 | P>S | No | EVA | |
| rs3388661742 | 363 | L>H | No | EVA |
No associated diseases with Q9JK48
Functions
14 GO annotations of cellular component
| Name | Definition |
|---|---|
| autophagosome membrane | The lipid bilayer surrounding an autophagosome, a double-membrane-bounded vesicle in which endogenous cellular material is sequestered. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early endosome | A membrane-bounded organelle that receives incoming material from primary endocytic vesicles that have been generated by clathrin-dependent and clathrin-independent endocytosis; vesicles fuse with the early endosome to deliver cargo for sorting into recycling or degradation pathways. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| mitochondrial envelope | The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space. |
| mitochondrial outer membrane | The outer, i.e. cytoplasm-facing, lipid bilayer of the mitochondrial envelope. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| fatty acid binding | Binding to a fatty acid, an aliphatic monocarboxylic acids liberated from naturally occurring fats and oils by hydrolysis. |
| identical protein binding | Binding to an identical protein or proteins. |
| lysophosphatidic acid acyltransferase activity | Catalysis of the transfer of acyl groups from an acyl-CoA to lysophosphatidic acid to form phosphatidic acid. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
25 GO annotations of biological process
| Name | Definition |
|---|---|
| 'de novo' post-translational protein folding | The process of assisting in the correct noncovalent folding of newly formed polypeptides or folding intermediates of polypeptides that have exited the ribosome and/or have been stabilized and transferred by other chaperone proteins. This process could involve several cycles of ATP hydrolysis. |
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| autophagic cell death | A form of programmed cell death that is accompanied by the formation of autophagosomes. Autophagic cell death is characterized by lack of chromatin condensation and massive vacuolization of the cytoplasm, with little or no uptake by phagocytic cells. |
| cellular response to amino acid starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of amino acids. |
| cellular response to glucose starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of glucose. |
| membrane fission | A process that is carried out at the cellular level which results in the separation of a single continuous membrane into two membranes. |
| membrane organization | A process which results in the assembly, arrangement of constituent parts, or disassembly of a membrane. A membrane is a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins. |
| mitochondrion organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a mitochondrion; includes mitochondrial morphogenesis and distribution, and replication of the mitochondrial genome as well as synthesis of new mitochondrial components. |
| negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | Any process that stops, prevents or reduces the frequency, rate or extent of intrinsic apoptotic signaling pathway by p53 class mediator. |
| phosphatidic acid biosynthetic process | The chemical reactions and pathways resulting in the formation of phosphatidic acid, any derivative of glycerol phosphate in which both the remaining hydroxyl groups of the glycerol moiety are esterified with fatty acids. |
| phospholipid biosynthetic process | The chemical reactions and pathways resulting in the formation of a phospholipid, a lipid containing phosphoric acid as a mono- or diester. |
| positive regulation of autophagosome assembly | Any process that activates or increases the frequency, rate or extent of autophagic vacuole assembly. |
| positive regulation of autophagy | Any process that activates, maintains or increases the rate of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| positive regulation of dendrite extension | Any process that activates or increases the frequency, rate or extent of dendrite extension. |
| positive regulation of dendrite morphogenesis | Any process that activates or increases the frequency, rate or extent of dendrite morphogenesis. |
| positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | Any process that activates or increases the frequency, rate or extent of intrinsic apoptotic signaling pathway by p53 class mediator. |
| positive regulation of membrane tubulation | Any process that activates or increases the frequency, rate or extent of membrane tubulation. |
| positive regulation of neurotrophin TRK receptor signaling pathway | Any process that activates or increases the frequency, rate or extent of the neurotrophin TRK receptor signaling pathway. |
| positive regulation of phosphatidylinositol 3-kinase activity | Any process that activates or increases the frequency, rate or extent of phosphatidylinositol 3-kinase activity. |
| positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | Any process that activates or increases the frequency, rate or extent of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway. |
| positive regulation of protein-containing complex assembly | Any process that activates or increases the frequency, rate or extent of protein complex assembly. |
| protein localization to vacuolar membrane | A process in which a protein is transported to, or maintained in, a location within a vacuolar membrane. |
| receptor catabolic process | The chemical reactions and pathways resulting in the breakdown of a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| regulation of cytokinesis | Any process that modulates the frequency, rate or extent of the division of the cytoplasm of a cell and its separation into two daughter cells. |
| regulation of early endosome to late endosome transport | Any process that modulates the frequency, rate or extent of early endosome to late endosome transport. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJA1 | SH3GL1 | Endophilin-A2 | Bos taurus (Bovine) | PR |
| Q8AXV1 | SH3GL2 | Endophilin-A1 | Gallus gallus (Chicken) | PR |
| Q9Y371 | SH3GLB1 | Endophilin-B1 | Homo sapiens (Human) | PR |
| Q99962 | SH3GL2 | Endophilin-A1 | Homo sapiens (Human) | PR |
| Q99961 | SH3GL1 | Endophilin-A2 | Homo sapiens (Human) | PR |
| Q99963 | SH3GL3 | Endophilin-A3 | Homo sapiens (Human) | PR |
| Q8BZT2 | Sh3rf2 | E3 ubiquitin-protein ligase SH3RF2 | Mus musculus (Mouse) | PR |
| Q62420 | Sh3gl2 | Endophilin-A1 | Mus musculus (Mouse) | PR |
| Q62419 | Sh3gl1 | Endophilin-A2 | Mus musculus (Mouse) | PR |
| Q62421 | Sh3gl3 | Endophilin-A3 | Mus musculus (Mouse) | PR |
| Q9JLQ0 | Cd2ap | CD2-associated protein | Mus musculus (Mouse) | SS |
| Q8R550 | Sh3kbp1 | SH3 domain-containing kinase-binding protein 1 | Mus musculus (Mouse) | SS |
| Q6AYE2 | Sh3glb1 | Endophilin-B1 | Rattus norvegicus (Rat) | PR |
| O35179 | Sh3gl2 | Endophilin-A1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNIMDFNVKK | LAADAGTFLS | RAVQFTEEKL | GQAEKTELDA | HLENLLSKAE | CTKIWTEKIM |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KQTEVLLQPN | PNARIEEFVY | EKLDRKAPSR | INNPELLGQY | MIDAGTEFGP | GTAYGNALIK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| CGETQKRIGT | ADRELIQTSA | LNFLTPLRNF | IEGDYKTIAK | ERKLLQNKRL | DLDAAKTRLK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KAKAAETKSS | SEQELRITQS | EFDRQAEITR | LLLEGISSTH | AHHLRCLNDF | VEAQMTYYAQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| CYQYMLDLQK | QLGSFPSNYL | SNNNQTSGTP | VPYALSNAIG | PSAQASTGSL | VITCPSNLND |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LKESSNNRKA | RVLYDYDAAN | STELSLLADE | VITVFSVVGM | DSDWLMGERG | NQKGKVPITY |
| LELLN |