Q9JJ25
Gene name |
Mefv |
Protein name |
Pyrin |
Names |
|
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
rno:58923 |
EC number |
|
Protein Class |
|
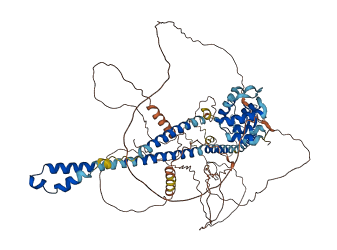
Descriptions
(Annotation from UniProt)
The B box-type zinc finger interacts, possibly intramolecularly, with the pyrin domain. This may be an autoinhibitory mechanism released by PSTPIP1 binding.
Autoinhibitory domains (AIDs)
Target domain |
1-92 (Pyrin domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for Q9JJ25
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9JJ25-F1 | Predicted | AlphaFoldDB |
No variants for Q9JJ25
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q9JJ25 | |||||
No associated diseases with Q9JJ25
Functions
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| autophagosome | A double-membrane-bounded compartment that engulfs endogenous cellular material as well as invading microorganisms to target them to the lytic vacuole/lysosome for degradation as part of macroautophagy. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| microtubule associated complex | Any multimeric complex connected to a microtubule. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| identical protein binding | Binding to an identical protein or proteins. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| zinc ion binding | Binding to a zinc ion (Zn). |
17 GO annotations of biological process
| Name | Definition |
|---|---|
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| negative regulation of cytokine production involved in inflammatory response | Any process that stops, prevents or reduces the frequency, rate or extent of cytokine production involved in inflammatory response. |
| negative regulation of inflammatory response | Any process that stops, prevents, or reduces the frequency, rate or extent of the inflammatory response. |
| negative regulation of interleukin-1 beta production | Any process that stops, prevents, or reduces the frequency, rate, or extent of interleukin-1 beta production. |
| negative regulation of interleukin-12 production | Any process that stops, prevents, or reduces the frequency, rate, or extent of interleukin-12 production. |
| negative regulation of macrophage inflammatory protein 1 alpha production | Any process that stops, prevents, or reduces the frequency, rate, or extent of production of macrophage inflammatory protein 1 alpha. |
| negative regulation of NLRP3 inflammasome complex assembly | Any process that stops, prevents or reduces the frequency, rate or extent of NLRP3 inflammasome complex assembly. |
| positive regulation of autophagy | Any process that activates, maintains or increases the rate of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| positive regulation of cysteine-type endopeptidase activity | Any process that activates or increases the frequency, rate or extent of cysteine-type endopeptidase activity. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| pyroptosome complex assembly | The aggregation, arrangement and bonding together of a set of components to form a pyroptosome complex. |
| regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| regulation of interleukin-1 beta production | Any process that modulates the frequency, rate, or extent of interleukin-1 beta production. |
| response to interferon-gamma | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interferon-gamma stimulus. Interferon-gamma is also known as type II interferon. |
| response to lipopolysaccharide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| response to silicon dioxide | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a silicon dioxide stimulus. |
46 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5E9G4 | TRIM10 | Tripartite motif-containing protein 10 | Bos taurus (Bovine) | PR |
| Q2T9Z0 | TRIM17 | E3 ubiquitin-protein ligase TRIM17 | Bos taurus (Bovine) | PR |
| E1BJS7 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Bos taurus (Bovine) | PR |
| Q7YRV4 | TRIM21 | E3 ubiquitin-protein ligase TRIM21 | Bos taurus (Bovine) | PR |
| Q1PRL4 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Gallus gallus (Chicken) | PR |
| Q7YR32 | TRIM10 | Tripartite motif-containing protein 10 | Pan troglodytes (Chimpanzee) | PR |
| Q9BTV5 | FSD1 | Fibronectin type III and SPRY domain-containing protein 1 | Homo sapiens (Human) | PR |
| Q9H2S5 | RNF39 | RING finger protein 39 | Homo sapiens (Human) | PR |
| P19474 | TRIM21 | E3 ubiquitin-protein ligase TRIM21 | Homo sapiens (Human) | PR |
| Q9UJV3 | MID2 | Probable E3 ubiquitin-protein ligase MID2 | Homo sapiens (Human) | PR |
| P29590 | PML | Protein PML | Homo sapiens (Human) | PR |
| Q9C029 | TRIM7 | E3 ubiquitin-protein ligase TRIM7 | Homo sapiens (Human) | PR |
| Q9NQ86 | TRIM36 | E3 ubiquitin-protein ligase TRIM36 | Homo sapiens (Human) | PR |
| Q86UV6 | TRIM74 | Tripartite motif-containing protein 74 | Homo sapiens (Human) | PR |
| Q9UPQ4 | TRIM35 | E3 ubiquitin-protein ligase TRIM35 | Homo sapiens (Human) | PR |
| Q6ZMU5 | TRIM72 | Tripartite motif-containing protein 72 | Homo sapiens (Human) | PR |
| Q86UV7 | TRIM73 | Tripartite motif-containing protein 73 | Homo sapiens (Human) | PR |
| Q8N9V2 | TRIML1 | Probable E3 ubiquitin-protein ligase TRIML1 | Homo sapiens (Human) | PR |
| Q86XT4 | TRIM50 | E3 ubiquitin-protein ligase TRIM50 | Homo sapiens (Human) | PR |
| Q5EBN2 | TRIM61 | Putative tripartite motif-containing protein 61 | Homo sapiens (Human) | PR |
| Q9BZY9 | TRIM31 | E3 ubiquitin-protein ligase TRIM31 | Homo sapiens (Human) | PR |
| Q2Q1W2 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Homo sapiens (Human) | PR |
| Q9BXM9 | FSD1L | FSD1-like protein | Homo sapiens (Human) | PR |
| O15553 | MEFV | Pyrin | Homo sapiens (Human) | SS |
| Q9WUH5 | Trim10 | Tripartite motif-containing protein 10 | Mus musculus (Mouse) | PR |
| Q8BZT2 | Sh3rf2 | E3 ubiquitin-protein ligase SH3RF2 | Mus musculus (Mouse) | PR |
| Q7TPM3 | Trim17 | E3 ubiquitin-protein ligase TRIM17 | Mus musculus (Mouse) | PR |
| Q1XH17 | Trim72 | Tripartite motif-containing protein 72 | Mus musculus (Mouse) | PR |
| Q60953 | Pml | Protein PML | Mus musculus (Mouse) | PR |
| Q99PQ1 | Trim12a | Tripartite motif-containing protein 12A | Mus musculus (Mouse) | PR |
| Q61510 | Trim25 | E3 ubiquitin/ISG15 ligase TRIM25 | Mus musculus (Mouse) | PR |
| Q810I2 | Trim50 | E3 ubiquitin-protein ligase TRIM50 | Mus musculus (Mouse) | PR |
| Q1PSW8 | Trim71 | E3 ubiquitin-protein ligase TRIM71 | Mus musculus (Mouse) | PR |
| Q3TL54 | Trim43a | Tripartite motif-containing protein 43A | Mus musculus (Mouse) | PR |
| P86449 | Trim43c | Tripartite motif-containing protein 43C | Mus musculus (Mouse) | PR |
| Q9JJ26 | Mefv | Pyrin | Mus musculus (Mouse) | SS |
| O77666 | TRIM26 | Tripartite motif-containing protein 26 | Sus scrofa (Pig) | PR |
| O19085 | TRIM10 | Tripartite motif-containing protein 10 | Sus scrofa (Pig) | PR |
| Q865W2 | TRIM50 | E3 ubiquitin-protein ligase TRIM50 | Sus scrofa (Pig) | PR |
| D3ZVM4 | Trim71 | E3 ubiquitin-protein ligase TRIM71 | Rattus norvegicus (Rat) | PR |
| Q920M2 | Rnf39 | RING finger protein 39 | Rattus norvegicus (Rat) | PR |
| A0JPQ4 | Trim72 | Tripartite motif-containing protein 72 | Rattus norvegicus (Rat) | PR |
| Q810I1 | Trim50 | E3 ubiquitin-protein ligase TRIM50 | Rattus norvegicus (Rat) | PR |
| F6QEU4 | trim71 | E3 ubiquitin-protein ligase TRIM71 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q640S6 | trim72 | Tripartite motif-containing protein 72 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| E7FAM5 | trim71 | E3 ubiquitin-protein ligase TRIM71 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MANTRVDHLL | NTLEELLPYE | LEKFKFKLHT | TSLEKGHSRI | PLSLVKMARP | IKLTRLLLTY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YGEEYAVRLT | LQILRATNQR | QLAEELHKAT | GPEHLTEENG | VGGSVQSSAE | NKDKGVKGSD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VPGEDEAQQN | DDESDILPPI | QAEVGKGPQK | KSLAKRKDQR | GPESLDSQTK | PGARSAAPLY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RRTLVTQSPG | DKENRAGAQP | QECQLCREAA | RSTAMSQGGE | RSRRLKCICL | QERSDPGVLK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LPLTQKKENP | QIQKLFRLKR | KQEMAVSFVR | EATLNGRTTG | TLEKGVGIPE | HSMMLDEETS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RNMSSKISLT | REKRCTASWT | ENGNGGPETP | ETLGETVSSI | LCDSCSPKVL | LSLGEKLAQT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PEDPASLGQA | ASKGRSRDKV | ACPLCHTQGE | LPAKACVQSS | CSCSVAPGDP | KASGRHSICF |
| 430 | 440 | 450 | 460 | 470 | 480 |
| QCQSSRAGKS | CEAQSPQFLP | QCPRHMKQVQ | LLFCEDHREP | ICLICRLSQE | HQGHRVRPIE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| EAALQYKEQI | RKQLERLREM | RGYVEEHKLP | ADKKAEDFLK | QTETQKQRIS | CPLEKLFQFL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EQQEQLFVTW | LQELVQTIGK | VRETYYTQVS | LLDKLIGELE | AKQDQPEWEL | MQDIGATLHR |
| 610 | 620 | 630 | 640 | 650 | 660 |
| AETMTASELL | GIPPGVKEKL | HLLYQKSKSA | EKNMQRFSEM | LGSEMAFSAS | DVATREGCRP |
| 670 | 680 | 690 | 700 | 710 | 720 |
| STTKAQALIP | TVHLKCDGAH | TQDFDVILCA | ELEAGGSEPQ | DYLHPSSAQD | TPELHEIHSQ |
| 730 | 740 | ||||
| NNKRKFKSFL | KWKPSFSRTD | RCLRTCW |