Q9HC98
Gene name |
NEK6 |
Protein name |
Serine/threonine-protein kinase Nek6 |
Names |
Never in mitosis A-related kinase 6, NimA-related protein kinase 6, Protein kinase SID6-1512 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:10783 |
EC number |
2.7.11.34: Protein-serine/threonine kinases |
Protein Class |
MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 20-RELATED (PTHR43289) |
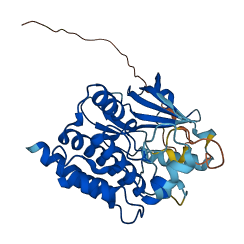
Descriptions
Nek6 is a NIMA-related kinase essential for proper mitotic spindle assembly and has a catalytic domain with a 30-40 amino acid N-terminal extension. Nek6 is subject to autoinhibition by the tyrosine motif, Tyr108. This is because the Y108A mutation is more active than wild-type Nek6. In addition, the activity of Nek6 is increased by interaction with the Nek9 noncatalytic C-terminal domain, which may release tyrosine from the autoinhibitory position.
Autoinhibitory domains (AIDs)
Target domain |
31-41 (N-terminal extension of kinase domain) |
Relief mechanism |
Partner binding |
Assay |
Structural analysis, Mutagenesis experiment |
Accessory elements
189-212 (Activation loop from InterPro)
Target domain |
45-310 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9HC98
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9HC98-F1 | Predicted | AlphaFoldDB |
202 variants for Q9HC98
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA374875763 rs1247048658 |
2 | A>V | No |
ClinGen gnomAD |
|
|
CA199686987 rs911139179 |
4 | Q>E | No |
ClinGen TOPMed |
|
|
CA5234091 rs779271535 |
4 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA374875781 rs1167516898 |
5 | P>L | No |
ClinGen gnomAD |
|
|
CA374875782 rs528727476 |
6 | G>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA5234093 rs528727476 |
6 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1365078463 CA374875792 |
7 | H>L | No |
ClinGen TOPMed gnomAD |
|
|
CA374875791 rs1365078463 |
7 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
CA374875801 rs1326135942 |
8 | M>I | No |
ClinGen gnomAD |
|
|
CA5234094 rs780230637 |
8 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA374875807 rs1331996591 |
9 | P>R | No |
ClinGen gnomAD |
|
|
rs377403028 CA5234095 |
10 | H>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1366906036 CA374875816 |
11 | G>R | No |
ClinGen gnomAD |
|
|
rs1588482166 CA374875835 |
13 | S>R | No |
ClinGen Ensembl |
|
|
CA374875840 rs1465974668 |
14 | S>C | No |
ClinGen gnomAD |
|
|
CA374875846 rs1454488440 |
15 | N>S | No |
ClinGen TOPMed |
|
|
CA5234096 rs145488745 |
16 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA374875873 rs1290364170 |
18 | C>Y | No |
ClinGen gnomAD |
|
|
rs747894737 CA5234098 |
21 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA374875936 rs867678055 |
22 | G>R | No |
ClinGen TOPMed |
|
|
CA199687059 rs867678055 |
22 | G>W | No |
ClinGen TOPMed |
|
|
CA374875951 rs1290547838 |
23 | P>L | No |
ClinGen gnomAD |
|
|
rs1446146786 CA374875953 |
24 | V>M | No |
ClinGen gnomAD |
|
|
CA5234099 rs769478376 |
25 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA199687064 rs140978005 |
26 | P>S | No |
ClinGen ESP gnomAD |
|
|
CA199687068 rs910941666 |
27 | P>L | No |
ClinGen gnomAD |
|
|
rs941098694 CA199687088 |
29 | P>S | No |
ClinGen Ensembl |
|
|
CA374876051 rs1384634882 |
30 | Q>R | No |
ClinGen gnomAD |
|
|
CA5234123 rs758964365 |
34 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA374877205 rs1256928848 |
34 | N>K | No |
ClinGen gnomAD |
|
|
CA5234124 rs769243779 |
35 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA5234125 rs774694008 |
35 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5234128 rs750746819 |
39 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1376354948 CA374877234 |
39 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA374877233 rs1376354948 |
39 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
CA374877238 rs1467238937 |
40 | C>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs763278157 CA5234129 |
41 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1409086630 CA374877248 |
42 | L>M | No |
ClinGen TOPMed gnomAD |
|
|
CA374877252 rs1433128929 |
42 | L>R | No |
ClinGen gnomAD |
|
|
rs1409086630 CA374877249 |
42 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA5234131 rs183673274 |
43 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs148262318 CA5234134 |
48 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs755980284 CA5234135 |
49 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs777558111 CA5234136 |
52 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA5234137 rs749069904 |
53 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1341208772 CA374877344 |
56 | F>Y | No |
ClinGen TOPMed |
|
|
rs745431350 CA5234140 |
58 | E>A | No |
ClinGen ExAC gnomAD |
|
|
CA5234139 rs778399540 |
58 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA199696983 rs923279115 |
63 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1588497542 CA374877391 |
63 | T>P | No |
ClinGen Ensembl |
|
|
CA374877409 rs1265646448 |
66 | L>M | No |
ClinGen gnomAD |
|
|
CA5234141 rs771504531 |
68 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374877426 rs1377794796 |
68 | R>S | No |
ClinGen TOPMed |
|
|
rs775022069 CA5234142 |
70 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374877478 rs1427488678 |
76 | V>A | No |
ClinGen gnomAD |
|
|
rs1372158895 CA374877475 |
76 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA374877474 rs1372158895 |
76 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA374877482 rs1370678576 |
77 | Q>* | No |
ClinGen gnomAD |
|
|
CA374877484 rs1429957019 |
77 | Q>R | No |
ClinGen TOPMed |
|
|
CA374877513 rs1386238966 |
79 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1332109613 CA374877532 |
82 | M>V | No |
ClinGen TOPMed |
|
|
rs779439151 CA5234159 |
84 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1386167089 CA374877565 |
86 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA199698616 rs987052518 |
88 | Q>R | No |
ClinGen Ensembl |
|
|
CA374877581 rs1588499962 |
89 | D>N | No |
ClinGen Ensembl |
|
|
rs183711992 CA199698654 |
92 | K>R | No |
ClinGen 1000Genomes gnomAD |
|
|
rs183711992 CA374877606 |
92 | K>T | No |
ClinGen 1000Genomes gnomAD |
|
|
rs1463842648 CA374877612 |
93 | E>* | No |
ClinGen gnomAD |
|
|
rs774713096 CA5234165 |
95 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs768754460 CA5234188 |
100 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA374869994 rs1209363432 |
101 | N>S | No |
ClinGen TOPMed |
|
|
rs1271939785 CA374870054 |
109 | L>W | No |
ClinGen gnomAD |
|
|
CA199658042 rs1031164724 |
111 | S>L | No |
ClinGen TOPMed |
|
|
CA374870074 rs1182869786 |
112 | F>C | No |
ClinGen gnomAD |
|
|
CA374870080 rs1267853445 |
113 | I>V | No |
ClinGen gnomAD |
|
|
rs370008827 CA5234193 |
114 | E>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs750249273 CA5234192 |
114 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs370008827 CA374870088 |
114 | E>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA374870093 rs1415316825 |
115 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1168037323 CA374870100 |
116 | N>D | No |
ClinGen gnomAD |
|
|
CA5234195 rs143094065 |
117 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5234197 rs780863742 |
119 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA374870173 rs1417581726 |
127 | A>T | No |
ClinGen TOPMed |
|
|
CA5234204 rs747235761 |
132 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs780356311 CA5234203 |
132 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374870223 rs1449600656 |
134 | I>N | No |
ClinGen gnomAD |
|
|
rs1223479281 CA374870228 |
135 | K>E | No |
ClinGen gnomAD |
|
|
rs868482996 CA199660864 |
136 | Y>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1472585159 CA374870530 |
138 | K>R | No |
ClinGen gnomAD |
|
|
rs1157394651 CA374870555 |
140 | Q>R | No |
ClinGen gnomAD |
|
|
CA199660870 rs759138170 |
142 | R>Q | No |
ClinGen TOPMed |
|
|
rs767329239 CA374870578 |
142 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs775228582 CA5234232 |
143 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA374870596 rs1454102379 |
144 | I>V | No |
ClinGen TOPMed |
|
|
CA5234233 rs368466543 |
145 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1395923597 CA374870621 |
146 | E>Q | No |
ClinGen TOPMed |
|
|
CA199660891 rs552985155 |
149 | V>A | No |
ClinGen 1000Genomes TOPMed |
|
|
rs1588517960 CA374870697 |
152 | Y>D | No |
ClinGen Ensembl |
|
|
CA374871106 rs1588517996 |
154 | V>A | No |
ClinGen Ensembl |
|
|
rs1407961158 CA374871100 |
154 | V>L | No |
ClinGen gnomAD |
|
|
CA374871099 rs1407961158 |
154 | V>M | No |
ClinGen gnomAD |
|
|
CA374871113 rs1289955664 |
155 | Q>* | No |
ClinGen gnomAD |
|
|
CA374871137 rs1483225181 |
157 | C>Y | No |
ClinGen TOPMed |
|
|
rs200565078 CA5234238 |
158 | S>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA199660913 rs879292381 |
159 | A>T | No |
ClinGen gnomAD |
|
|
rs751958560 CA5234241 |
160 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5234240 rs751958560 |
160 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5234243 rs748409240 |
165 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs369950028 CA5234244 |
166 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1420088183 CA374871249 |
166 | R>H | No |
ClinGen gnomAD |
|
|
CA374871244 rs369950028 |
166 | R>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs777970915 CA5234245 |
167 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1461941868 CA374871256 |
167 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA5234247 rs771033424 |
171 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA374871305 rs1369451980 |
171 | R>Q | No |
ClinGen gnomAD |
|
|
CA5234282 rs760926146 |
175 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs553709380 CA374871493 |
177 | N>K | No |
ClinGen gnomAD |
|
|
CA374871497 rs1482758201 |
178 | V>M | No |
ClinGen gnomAD |
|
|
rs1162626313 CA374871542 |
182 | A>T | No |
ClinGen gnomAD |
|
|
CA5234284 rs754146573 |
183 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5234286 rs779146612 |
185 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5234288 rs758428107 |
186 | V>G | No |
ClinGen ExAC gnomAD |
|
|
CA5234287 rs775352569 |
186 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs779821672 CA374871617 |
189 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779821672 CA5234289 |
189 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5234291 rs768356782 |
195 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA374871678 rs768356782 |
195 | R>S | No |
ClinGen ExAC gnomAD |
|
|
rs200086787 CA199661854 |
196 | F>C | No |
ClinGen Ensembl |
|
|
rs201207004 CA199661861 |
197 | F>L | No |
ClinGen gnomAD |
|
|
rs747816154 CA5234293 |
200 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs1588519923 CA374871751 |
201 | T>P | No |
ClinGen Ensembl |
|
|
rs1588519942 CA374871761 |
202 | T>P | No |
ClinGen Ensembl |
|
|
rs772780677 CA5234296 |
203 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs751648398 CA5234330 |
215 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA5234331 rs555077914 |
216 | P>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA5234332 rs767414917 |
216 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5234334 rs755926840 |
220 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA5234336 rs543844372 |
224 | Y>* | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA199668393 rs201833354 |
230 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs1042477188 CA199668386 |
230 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA5234339 rs779050538 |
232 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA374873314 rs10760354 |
233 | L>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA5234341 rs771906905 |
237 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA374873377 rs1288821162 |
239 | E>K | No |
ClinGen gnomAD |
|
|
CA374874481 rs1190475979 |
242 | A>T | No |
ClinGen gnomAD |
|
|
rs1419093540 CA374874497 |
243 | L>F | No |
ClinGen gnomAD |
|
|
CA5234366 rs151138282 |
252 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs919717766 CA199675081 |
252 | M>K | No |
ClinGen Ensembl |
|
|
CA5234365 rs749637388 |
252 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1338625653 CA374874674 |
255 | F>L | No |
ClinGen gnomAD |
|
|
CA374874669 rs1588554508 |
255 | F>S | No |
ClinGen Ensembl |
|
|
rs774656051 CA5234367 |
256 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774656051 CA5234368 |
256 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772144294 CA5234369 |
259 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA5234372 rs764086271 |
262 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA5234373 rs753614385 |
263 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374874733 rs1315306769 |
264 | C>F | No |
ClinGen TOPMed |
|
|
CA5234374 rs761645776 |
264 | C>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5234375 rs764888197 |
265 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA5234376 rs750118016 |
266 | Y>F | No |
ClinGen ExAC |
|
|
CA374874746 rs750118016 |
266 | Y>S | No |
ClinGen ExAC |
|
|
rs779720887 CA5234379 |
267 | P>H | No |
ClinGen ExAC gnomAD |
|
|
CA5234382 rs778322409 |
268 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1564666860 CA374874774 |
271 | G>E | No |
ClinGen Ensembl |
|
|
CA5234385 rs779084313 COSM1597960 |
271 | G>R | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1564666865 CA374874777 |
272 | E>K | No |
ClinGen Ensembl |
|
|
CA374874794 rs1301876690 |
274 | Y>H | No |
ClinGen TOPMed |
|
|
CA5234386 rs746106299 |
275 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
rs144052872 CA5234388 |
276 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs144052872 CA374874806 |
276 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs760711060 CA5234389 |
277 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs768520677 CA5234390 |
277 | K>M | No |
ClinGen ExAC gnomAD |
|
|
rs200398623 CA5234408 |
279 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1469571292 CA374874934 |
280 | E>* | No |
ClinGen gnomAD |
|
|
CA374874941 rs1193711970 |
281 | L>V | No |
ClinGen gnomAD |
|
|
rs758656747 CA5234409 |
283 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA199677586 rs201113183 |
284 | M>T | No |
ClinGen Ensembl |
|
|
CA5234410 rs780223937 |
284 | M>V | No |
ClinGen ExAC |
|
|
CA5234411 rs747180914 |
285 | C>S | No |
ClinGen ExAC gnomAD |
|
|
CA374874968 rs747180914 |
285 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs776507491 CA5234413 |
287 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs748129024 CA5234415 |
289 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374875002 rs1248451252 |
290 | P>A | No |
ClinGen TOPMed |
|
|
CA5234417 rs146443565 |
291 | H>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA5234418 rs762820475 |
292 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1298408686 CA374875034 |
295 | D>N | No |
ClinGen gnomAD |
|
|
rs1271742411 CA374875053 |
297 | G>A | No |
ClinGen gnomAD |
|
|
rs774178334 CA5234420 |
297 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs200235098 CA374875062 |
299 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5234422 rs200235098 |
299 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs542509176 CA5234424 |
300 | H>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs376476122 CA5234425 |
301 | Q>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA374875084 rs1423213989 |
302 | V>A | No |
ClinGen gnomAD |
|
|
CA199677681 rs954971092 |
302 | V>L | No |
ClinGen TOPMed |
|
|
rs1414669747 CA374875122 |
307 | H>R | No |
ClinGen TOPMed |
|
|
CA199677696 rs915086313 |
309 | W>* | No |
ClinGen Ensembl |
|
|
rs1395201958 CA374875134 |
309 | W>G | No |
ClinGen TOPMed |
|
|
CA374875142 rs1165927187 |
310 | M>L | No |
ClinGen gnomAD |
|
|
CA374875162 rs1344640329 |
312 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA374875168 rs1451744753 |
313 | T>I | No |
ClinGen gnomAD |
|
|
rs1290755558 CA374875170 |
314 | T>R | No |
ClinGen gnomAD |
|
|
CA374875175 rs1363269245 |
314 | T>W | No |
ClinGen gnomAD |
No associated diseases with Q9HC98
6 regional properties for Q9HC98
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 45 - 310 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 122 - 135 | IPR001245-1 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 162 - 180 | IPR001245-2 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 228 - 250 | IPR001245-3 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 274 - 296 | IPR001245-4 |
| active_site | Serine/threonine-protein kinase, active site | 168 - 180 | IPR008271 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.34 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR43289 | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 20-RELATED |
| PANTHER Subfamily | PTHR43289:SF13 | SERINE_THREONINE-PROTEIN KINASE NEK6 |
| PANTHER Protein Class |
non-receptor serine/threonine protein kinase
protein modifying enzyme |
|
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| centriolar satellite | A small (70-100 nm) cytoplasmic granule that contains a number of centrosomal proteins; centriolar satellites traffic toward microtubule minus ends and are enriched near the centrosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| nuclear speck | A discrete extra-nucleolar subnuclear domain, 20-50 in number, in which splicing factors are seen to be localized by immunofluorescence microscopy. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| spindle pole | Either of the ends of a spindle, where spindle microtubules are organized; usually contains a microtubule organizing center and accessory molecules, spindle microtubules and astral microtubules. |
10 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| kinesin binding | Interacting selectively and non-covalently and stoichiometrically with kinesin, a member of a superfamily of microtubule-based motor proteins that perform force-generating tasks such as organelle transport and chromosome segregation. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| transcription corepressor binding | Binding to a transcription corepressor, a protein involved in negative regulation of transcription via protein-protein interactions with transcription factors and other proteins that negatively regulate transcription. Transcription corepressors do not bind DNA directly, but rather mediate protein-protein interactions between repressing transcription factors and the basal transcription machinery. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| chromosome segregation | The process in which genetic material, in the form of chromosomes, is organized into specific structures and then physically separated and apportioned to two or more sets. In eukaryotes, chromosome segregation begins with the condensation of chromosomes, includes chromosome separation, and ends when chromosomes have completed movement to the spindle poles. |
| mitotic nuclear membrane disassembly | The mitotic cell cycle process in which the controlled partial or complete breakdown of the nuclear membranes during occurs during mitosis. |
| mitotic spindle organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the microtubule spindle during a mitotic cell cycle. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of I-kappaB kinase/NF-kappaB signaling | Any process that activates or increases the frequency, rate or extent of I-kappaB kinase/NF-kappaB signaling. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of cellular senescence | Any process that modulates the frequency, rate or extent of cellular senescence. |
| regulation of mitotic cell cycle | Any process that modulates the rate or extent of progress through the mitotic cell cycle. |
| regulation of mitotic metaphase/anaphase transition | Any process that modulates the frequency, rate or extent of the cell cycle process in which a cell progresses from metaphase to anaphase during mitosis, triggered by the activation of the anaphase promoting complex by Cdc20/Sleepy homolog which results in the degradation of Securin. |
| spindle assembly | The aggregation, arrangement and bonding together of a set of components to form the spindle, the array of microtubules and associated molecules that serves to move duplicated chromosomes apart. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8TDX7 | NEK7 | Serine/threonine-protein kinase Nek7 | Homo sapiens (Human) | EV |
| Q00526 | CDK3 | Cyclin-dependent kinase 3 | Homo sapiens (Human) | PR |
| Q9ES74 | Nek7 | Serine/threonine-protein kinase Nek7 | Mus musculus (Mouse) | SS |
| Q9ES70 | Nek6 | Serine/threonine-protein kinase Nek6 | Mus musculus (Mouse) | SS |
| A2BD05 | NEK6 | Serine/threonine-protein kinase Nek6 | Sus scrofa (Pig) | SS |
| D3ZBE5 | Nek7 | Serine/threonine-protein kinase Nek7 | Rattus norvegicus (Rat) | SS |
| P59895 | Nek6 | Serine/threonine-protein kinase Nek6 | Rattus norvegicus (Rat) | SS |
| G5EFM9 | nekl-3 | Serine/threonine-protein kinase nekl-3 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAGQPGHMPH | GGSSNNLCHT | LGPVHPPDPQ | RHPNTLSFRC | SLADFQIEKK | IGRGQFSEVY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KATCLLDRKT | VALKKVQIFE | MMDAKARQDC | VKEIGLLKQL | NHPNIIKYLD | SFIEDNELNI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VLELADAGDL | SQMIKYFKKQ | KRLIPERTVW | KYFVQLCSAV | EHMHSRRVMH | RDIKPANVFI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TATGVVKLGD | LGLGRFFSSE | TTAAHSLVGT | PYYMSPERIH | ENGYNFKSDI | WSLGCLLYEM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AALQSPFYGD | KMNLFSLCQK | IEQCDYPPLP | GEHYSEKLRE | LVSMCICPDP | HQRPDIGYVH |
| 310 | |||||
| QVAKQMHIWM | SST |