Q9HBU1
Gene name |
BARX1 |
Protein name |
Homeobox protein BarH-like 1 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:56033 |
EC number |
|
Protein Class |
|
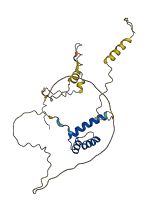
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q9HBU1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2DMT | NMR | - | A | 133-199 | PDB |
| AF-Q9HBU1-F1 | Predicted | AlphaFoldDB |
209 variants for Q9HBU1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1587810156 CA374334954 |
3 | R>W | No |
ClinGen Ensembl |
|
|
rs895896348 CA196447569 |
4 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA374334940 rs1304665974 |
5 | G>V | No |
ClinGen gnomAD |
|
|
rs1483686725 CA374334939 |
6 | E>K | No |
ClinGen TOPMed |
|
|
rs774263956 CA5133429 |
7 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM3395936 CA374334931 rs1190383513 |
7 | P>S | pancreas [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA374334911 rs1262291713 |
10 | A>E | No |
ClinGen gnomAD |
|
| TCGA novel | 10 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA374334892 rs1356935133 |
13 | G>C | No |
ClinGen gnomAD |
|
|
rs1356935133 CA374334894 |
13 | G>S | No |
ClinGen gnomAD |
|
|
rs770854053 CA5133428 |
14 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA374334879 rs1246522511 |
15 | P>R | No |
ClinGen gnomAD |
|
|
rs867917015 CA196447556 |
16 | E>* | No |
ClinGen Ensembl |
|
|
CA374334867 rs1338964571 |
17 | G>D | No |
ClinGen gnomAD |
|
|
rs1412540917 CA374334860 |
18 | C>S | No |
ClinGen TOPMed |
|
|
CA196447550 rs934796935 |
19 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
CA374334856 rs934796935 |
19 | A>T | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 19 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA374334848 rs1448262683 |
20 | D>A | No |
ClinGen TOPMed |
|
|
CA196447546 rs924748384 |
22 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA374334834 rs1314290662 |
22 | R>W | No |
ClinGen TOPMed |
|
|
CA374334821 rs1288504701 |
24 | H>R | No |
ClinGen TOPMed |
|
|
CA374334817 rs1353455167 |
25 | R>C | No |
ClinGen TOPMed |
|
|
CA374334814 rs1218028254 |
25 | R>H | No |
ClinGen TOPMed |
|
|
rs1327136481 CA374334800 |
27 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
CA196447542 rs866099103 |
28 | S>I | No |
ClinGen Ensembl |
|
|
rs1303027019 CA374334782 |
30 | M>L | No |
ClinGen gnomAD |
|
|
rs1253384506 CA374334772 |
31 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1201657492 CA374334732 |
36 | T>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
| TCGA novel | 37 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1182986365 CA374334713 |
39 | P>L | No |
ClinGen TOPMed |
|
|
rs1418324655 CA374334709 |
40 | G>E | No |
ClinGen TOPMed gnomAD |
|
|
rs781194809 CA5133426 |
40 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1476079637 CA374334695 |
42 | K>M | No |
ClinGen TOPMed gnomAD |
|
|
CA374334693 rs1373164993 |
42 | K>N | No |
ClinGen gnomAD |
|
|
rs1476079637 CA374334696 |
42 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA374334683 rs1489837185 |
44 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
CA196447525 rs560200241 |
44 | A>T | No |
ClinGen 1000Genomes gnomAD |
|
|
rs1489837185 CA374334682 |
44 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1175286609 CA374334678 |
45 | A>E | No |
ClinGen TOPMed |
|
|
rs1271031550 CA374334681 |
45 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA374334671 rs1563990316 |
46 | P>H | No |
ClinGen Ensembl |
|
|
rs1437054497 CA374334669 |
47 | A>T | No |
ClinGen gnomAD |
|
|
rs191789925 VAR_010927 CA5133425 |
48 | A>T | No |
ClinGen UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
CA374334656 rs1216574487 |
49 | A>T | No |
ClinGen gnomAD |
|
|
rs1340093500 CA374334652 |
49 | A>V | No |
ClinGen gnomAD |
|
|
rs576837358 CA5133424 |
51 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA374334643 rs1347925732 |
51 | A>V | No |
ClinGen gnomAD |
|
|
rs1441897044 CA374334635 |
52 | A>V | No |
ClinGen gnomAD |
|
|
CA374334624 rs1353143378 |
54 | G>A | No |
ClinGen gnomAD |
|
|
rs758702906 CA5133422 |
56 | L>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1400661440 CA374334601 |
58 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA374334597 rs1587810002 |
59 | F>L | No |
ClinGen Ensembl |
|
|
rs1273016991 CA374334589 |
60 | G>S | No |
ClinGen TOPMed |
|
|
rs1479197274 CA374334580 |
61 | V>E | No |
ClinGen TOPMed gnomAD |
|
|
CA374334578 rs1479197274 |
61 | V>G | No |
ClinGen TOPMed gnomAD |
|
|
CA196447495 rs964462092 |
62 | Q>R | No |
ClinGen TOPMed |
|
|
rs1200103921 CA374334566 |
63 | A>G | No |
ClinGen gnomAD |
|
|
rs941962960 CA374334551 |
66 | A>E | No |
ClinGen gnomAD |
|
|
CA196447492 rs941962960 |
66 | A>G | No |
ClinGen gnomAD |
|
|
CA374334544 rs1449899122 |
67 | A>V | No |
ClinGen TOPMed |
|
|
CA374334541 rs1563990263 |
68 | R>Q | No |
ClinGen Ensembl |
|
| TCGA novel | 69 | P>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1198280925 CA374334510 |
72 | S>R | No |
ClinGen TOPMed |
|
|
rs773164189 CA5133406 |
75 | A>D | No |
ClinGen ExAC gnomAD |
|
|
rs773164189 CA374334479 |
75 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA374334477 rs1313862806 |
76 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA374334463 rs1302918191 |
78 | K>R | No |
ClinGen gnomAD |
|
|
rs1489234016 CA374334451 |
80 | E>* | No |
ClinGen gnomAD |
|
|
rs1489234016 CA374334453 |
80 | E>K | No |
ClinGen gnomAD |
|
|
rs867705180 CA196446454 |
81 | Q>* | No |
ClinGen Ensembl |
|
|
rs865878957 CA196446450 |
84 | V>L | No |
ClinGen Ensembl |
|
|
CA374334414 rs1161255040 |
85 | F>L | No |
ClinGen gnomAD |
|
|
rs1171309735 CA374334412 |
86 | K>E | No |
ClinGen TOPMed |
|
|
CA5133403 rs747038748 |
86 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA374334397 rs1587809079 |
88 | P>A | No |
ClinGen Ensembl |
|
|
CA5133402 rs780245526 |
88 | P>Q | No |
ClinGen ExAC gnomAD |
|
|
rs541371621 CA374334392 |
89 | L>V | No |
ClinGen TOPMed |
|
|
rs746115723 CA5133400 |
92 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374334370 rs1185977992 |
93 | G>C | No |
ClinGen gnomAD |
|
|
rs1484955097 CA374334369 |
93 | G>D | No |
ClinGen gnomAD |
|
|
rs1185977992 CA374334371 |
93 | G>R | No |
ClinGen gnomAD |
|
|
CA5133399 rs549526966 |
94 | C>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs532945486 CA5133398 |
95 | S>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA374334358 rs1442164135 |
95 | S>P | No |
ClinGen TOPMed |
|
|
rs1343899 CA196446417 |
96 | G>R | No |
ClinGen Ensembl |
|
|
CA374334346 rs1379510866 |
97 | L>P | No |
ClinGen TOPMed |
|
|
CA5133396 rs778400548 |
98 | S>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA196446406 rs778400548 |
98 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1361139386 CA374334339 |
98 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1313049531 CA374334333 |
99 | S>F | No |
ClinGen gnomAD |
|
|
CA5133395 rs756740714 |
100 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1213446917 CA374334330 |
100 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 101 | L>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs45527632 CA196446393 |
103 | A>T | No |
ClinGen Ensembl |
|
|
CA374334308 rs1245867728 |
104 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
rs767061973 CA5133393 |
105 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs544075964 CA5133392 |
106 | P>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA5133391 rs544075964 |
106 | P>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA5133389 CA374334295 rs531602020 |
107 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs531602020 CA5133390 |
107 | G>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1421759690 CA374334280 |
110 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA374334273 rs201946673 |
111 | A>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA5133387 rs201946673 |
111 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1044700919 CA196446345 |
112 | A>T | No |
ClinGen TOPMed |
|
|
CA374334266 rs1267457561 |
112 | A>V | No |
ClinGen gnomAD |
|
|
rs1403090266 CA374334257 |
114 | A>T | No |
ClinGen TOPMed |
|
|
CA5133386 rs761812989 |
116 | H>P | No |
ClinGen ExAC gnomAD |
|
|
rs772214631 CA5133384 |
117 | L>M | No |
ClinGen ExAC gnomAD |
|
|
rs1218772762 CA374334229 |
118 | P>L | No |
ClinGen gnomAD |
|
|
CA5133381 rs771325755 |
121 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1391213209 CA374334202 |
122 | Q>H | No |
ClinGen gnomAD |
|
|
rs373384950 CA196446326 |
123 | L>F | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1460666524 CA374334192 COSM1184445 |
124 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA5133380 rs369928804 |
129 | A>S | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1427777963 CA374334154 |
130 | A>E | No |
ClinGen gnomAD |
|
|
rs868608877 CA374334148 |
131 | G>D | No |
ClinGen gnomAD |
|
|
CA196446306 rs868608877 |
131 | G>V | No |
ClinGen gnomAD |
|
|
rs778351778 CA5133379 |
132 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs770498173 CA196446301 |
134 | E>D | No |
ClinGen Ensembl |
|
|
rs748707341 CA5133377 |
136 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs780522907 CA5133376 |
137 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs780522907 CA374334114 |
137 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1198689451 CA374334099 |
139 | A>V | No |
ClinGen gnomAD |
|
|
CA5133375 rs754547962 |
140 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA374334080 rs1261122752 |
142 | G>E | No |
ClinGen gnomAD |
|
| TCGA novel | 142 | G>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA374334074 rs1331406485 |
143 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs866954519 CA196446272 |
145 | S>I | No |
ClinGen gnomAD |
|
|
rs866954519 CA374334063 |
145 | S>N | No |
ClinGen gnomAD |
|
|
rs1389519411 CA374334055 |
146 | R>H | No |
ClinGen gnomAD |
|
|
CA374334027 rs1332350470 |
151 | E>K | No |
ClinGen gnomAD |
|
|
CA374334012 rs1179141312 |
153 | Q>* | No |
ClinGen gnomAD |
|
|
rs1466998124 CA374334007 |
153 | Q>H | No |
ClinGen gnomAD |
|
| TCGA novel | 158 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs765995311 CA5133373 |
162 | E>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374333929 rs142710643 |
164 | Q>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1239576202 CA374333936 |
164 | Q>K | No |
ClinGen gnomAD |
|
|
rs750149824 CA5133371 |
169 | T>K | No |
ClinGen ExAC gnomAD |
|
|
rs750149824 COSM1111311 CA374333896 |
169 | T>M | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs531210852 CA196446241 |
170 | P>L | No |
ClinGen gnomAD |
|
|
rs778532537 CA5133352 |
173 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs1231334745 CA374333848 |
175 | L>I | No |
ClinGen TOPMed |
|
|
CA5133349 rs556916619 |
181 | L>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA5133348 rs201922522 |
184 | L>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA374333784 rs1263813289 |
185 | Q>* | No |
ClinGen gnomAD |
|
| TCGA novel | 187 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5133347 rs751625001 |
193 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA196446152 rs904775426 |
194 | R>M | No |
ClinGen Ensembl |
|
|
CA374333713 rs1198131040 |
195 | M>L | No |
ClinGen gnomAD |
|
|
rs1382974696 CA374333672 |
200 | I>V | No |
ClinGen gnomAD |
|
|
rs745365151 CA5133318 |
201 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs557180708 CA196445848 |
203 | Q>R | No |
ClinGen gnomAD |
|
|
CA374333635 rs1587808587 |
204 | G>C | No |
ClinGen Ensembl |
|
|
rs770548524 CA5133316 |
204 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA374333627 rs1175534327 |
205 | G>D | No |
ClinGen gnomAD |
|
|
rs1400402143 CA374333630 |
205 | G>S | No |
ClinGen gnomAD |
|
|
CA5133315 rs748908921 |
206 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs957172729 CA196445824 |
208 | E>A | No |
ClinGen TOPMed gnomAD |
|
|
rs957172729 CA374333611 |
208 | E>G | No |
ClinGen TOPMed gnomAD |
|
|
CA374333596 rs1180771426 |
210 | P>L | No |
ClinGen gnomAD |
|
|
CA5133314 rs149189558 |
211 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1239629658 CA374333594 |
211 | T>P | No |
ClinGen TOPMed |
|
| TCGA novel | 215 | G>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA374333560 rs1235663514 |
216 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1279996964 CA374333561 |
216 | R>W | No |
ClinGen gnomAD |
|
| TCGA novel | 217 | P>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs948370232 CA196445817 |
218 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA5133310 rs755024473 |
220 | N>I | No |
ClinGen ExAC gnomAD |
|
|
CA5133309 rs145379597 |
222 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA374333517 rs1157606968 |
223 | P>S | No |
ClinGen TOPMed |
|
|
rs992336948 CA196445787 |
224 | T>A | No |
ClinGen gnomAD |
|
|
rs757450124 CA5133307 |
224 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5133308 rs757450124 |
224 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746558122 CA196445762 |
225 | S>R | No |
ClinGen Ensembl |
|
|
CA196445760 rs961583468 |
226 | E>Q | No |
ClinGen TOPMed |
|
|
CA374333493 rs1324622909 |
227 | Q>K | No |
ClinGen gnomAD |
|
|
rs201122153 CA5133305 |
228 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs201122153 CA5133306 |
228 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 229 | T>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs560651229 CA5133304 |
230 | E>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA196445750 rs972503300 |
233 | R>C | No |
ClinGen TOPMed |
|
|
CA374333449 rs1378266524 |
233 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
CA374333453 rs972503300 |
233 | R>S | No |
ClinGen TOPMed |
|
|
CA374333446 rs1446509008 |
234 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1181808628 CA374333434 |
236 | D>N | No |
ClinGen gnomAD |
|
| TCGA novel | 238 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5133300 rs773850927 |
238 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1215715737 CA374333411 |
239 | K>* | No |
ClinGen gnomAD |
|
|
CA374333401 rs1288596242 |
240 | P>L | No |
ClinGen gnomAD |
|
|
rs770372550 CA5133299 |
240 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1313975275 CA374333398 |
241 | A>E | No |
ClinGen TOPMed gnomAD |
|
|
rs758410858 CA196445734 |
241 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1313975275 CA374333396 |
241 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1587808480 CA374333382 |
243 | V>G | No |
ClinGen Ensembl |
|
|
CA374333385 rs1226080454 |
243 | V>L | No |
ClinGen gnomAD |
|
|
CA374333378 rs1239139496 |
244 | P>L | No |
ClinGen TOPMed |
|
|
CA374333367 rs1429948249 |
246 | E>* | No |
ClinGen gnomAD |
|
|
CA374333357 rs1423783980 |
247 | P>R | No |
ClinGen gnomAD |
|
|
CA374333354 rs1358624725 |
248 | S>G | No |
ClinGen gnomAD |
|
|
rs772711999 CA5133297 |
248 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA374333346 rs1456235522 |
249 | D>H | No |
ClinGen TOPMed |
|
|
CA374333347 rs1456235522 |
249 | D>N | No |
ClinGen TOPMed |
|
|
CA374333348 rs1456235522 |
249 | D>Y | No |
ClinGen TOPMed |
|
|
CA374333334 rs1365144935 |
250 | R>S | No |
ClinGen gnomAD |
|
|
CA374333339 rs1171882411 |
250 | R>W | No |
ClinGen gnomAD |
|
|
rs374624014 CA196445726 |
252 | R>H | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs374624014 CA196445722 |
252 | R>L | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs199527359 CA5133294 |
253 | E>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
No associated diseases with Q9HBU1
5 regional properties for Q9HBU1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Helix-turn-helix motif | 171 - 196 | IPR000047 |
| domain | Homeobox domain | 140 - 204 | IPR001356 |
| conserved_site | Homeobox, conserved site | 175 - 198 | IPR017970 |
| domain | Homeobox domain, metazoa | 164 - 175 | IPR020479-1 |
| domain | Homeobox domain, metazoa | 179 - 198 | IPR020479-2 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| anterior/posterior pattern specification | The regionalization process in which specific areas of cell differentiation are determined along the anterior-posterior axis. The anterior-posterior axis is defined by a line that runs from the head or mouth of an organism to the tail or opposite end of the organism. |
| digestive system development | The process whose specific outcome is the progression of the digestive system over time, from its formation to the mature structure. The digestive system is the entire structure in which digestion takes place. Digestion is all of the physical, chemical, and biochemical processes carried out by multicellular organisms to break down ingested nutrients into components that may be easily absorbed and directed into metabolism. |
| endothelial cell differentiation | The process in which a mesodermal, bone marrow or neural crest cell acquires specialized features of an endothelial cell, a thin flattened cell. A layer of such cells lines the inside surfaces of body cavities, blood vessels, and lymph vessels, making up the endothelium. |
| negative regulation of Wnt signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of the Wnt signaling pathway. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| spleen development | The process whose specific outcome is the progression of the spleen over time, from its formation to the mature structure. The spleen is a large vascular lymphatic organ composed of white and red pulp, involved both in hemopoietic and immune system functions. |
| Wnt signaling pathway | The series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell and ending with a change in cell state. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9DED6 | BARX1B | Homeobox protein BarH-like 1b | Gallus gallus (Chicken) | PR |
| Q6RFL5 | BSX | Brain-specific homeobox protein homolog | Gallus gallus (Chicken) | PR |
| Q9W6D8 | BARX1 | Homeobox protein BarH-like 1 | Gallus gallus (Chicken) | PR |
| Q9UMQ3 | BARX2 | Homeobox protein BarH-like 2 | Homo sapiens (Human) | PR |
| Q3C1V8 | BSX | Brain-specific homeobox protein homolog | Homo sapiens (Human) | PR |
| Q9ER42 | Barx1 | Homeobox protein BarH-like 1 | Mus musculus (Mouse) | PR |
| Q810B3 | Bsx | Brain-specific homeobox protein homolog | Mus musculus (Mouse) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MQRPGEPGAA | RFGPPEGCAD | HRPHRYRSFM | IEEILTEPPG | PKGAAPAAAA | AAAGELLKFG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VQALLAARPF | HSHLAVLKAE | QAAVFKFPLA | PLGCSGLSSA | LLAAGPGLPG | AAGAPHLPLE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LQLRGKLEAA | GPGEPGTKAK | KGRRSRTVFT | ELQLMGLEKR | FEKQKYLSTP | DRIDLAESLG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LSQLQVKTWY | QNRRMKWKKI | VLQGGGLESP | TKPKGRPKKN | SIPTSEQLTE | QERAKDAEKP |
| 250 | |||||
| AEVPGEPSDR | SRED |