Q9H8T0
Gene name |
AKTIP |
Protein name |
AKT-interacting protein |
Names |
Ft1, Fused toes protein homolog |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:64400 |
EC number |
|
Protein Class |
|
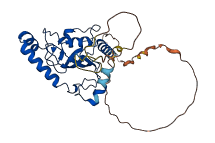
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9H8T0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9H8T0-F1 | Predicted | AlphaFoldDB |
178 variants for Q9H8T0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA395922361 rs1364147270 |
8 | S>F | No |
ClinGen gnomAD |
|
|
CA395922322 rs1364897796 |
12 | V>G | No |
ClinGen gnomAD |
|
|
CA8057095 COSM1378340 rs779924211 |
13 | R>C | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA8057094 rs769566710 |
13 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs1371166754 CA395922308 |
14 | K>R | No |
ClinGen gnomAD |
|
|
rs764155782 CA8057078 |
15 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1429434344 CA395922192 |
16 | S>F | No |
ClinGen gnomAD |
|
|
CA8057076 rs775441301 |
20 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs919044600 CA281365467 |
20 | E>K | No |
ClinGen Ensembl |
|
|
CA395922162 rs1250209537 |
21 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
rs61734118 CA8057075 |
21 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA395922151 rs1481856382 |
22 | T>I | No |
ClinGen gnomAD |
|
|
CA281365457 rs973534183 |
25 | G>R | No |
ClinGen Ensembl |
|
|
CA395922123 rs776539900 |
27 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8057072 rs776539900 |
27 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1225219264 CA395922100 |
30 | S>I | No |
ClinGen gnomAD |
|
|
rs770900884 CA8057071 |
31 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1428326756 CA395922090 |
32 | P>S | No |
ClinGen TOPMed |
|
|
rs141560947 CA8057070 |
33 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM971479 CA8057069 rs141560947 |
33 | R>Q | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs747621138 CA8057067 |
37 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs780558074 CA8057066 |
37 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756609255 CA8057065 |
42 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
rs986386188 CA281365403 |
43 | I>T | No |
ClinGen Ensembl |
|
| TCGA novel | 46 | N>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8057063 rs767844319 |
50 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs199669647 CA281365360 COSM3932365 |
51 | T>A | urinary_tract [Cosmic] | No |
ClinGen cosmic curated 1000Genomes |
|
rs751715236 CA8057061 |
53 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs533946422 CA8057062 |
53 | P>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs764315195 CA8057060 |
54 | T>K | No |
ClinGen ExAC gnomAD |
|
|
rs199548307 CA281365344 |
55 | S>C | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA395921951 rs1437597313 |
55 | S>P | No |
ClinGen TOPMed |
|
|
rs775464421 CA8057058 |
56 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8057056 rs759373692 |
58 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA8057055 rs759373692 |
58 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA8057053 rs770843306 |
60 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs908389472 CA281365313 |
62 | S>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1046908213 CA281365299 |
62 | S>L | No |
ClinGen TOPMed |
|
|
CA281365290 rs1021665141 |
63 | T>A | No |
ClinGen TOPMed |
|
|
rs1021665141 CA281365289 |
63 | T>S | No |
ClinGen TOPMed |
|
|
rs746802147 CA8057052 |
64 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs771641760 CA8057050 |
66 | T>M | No |
ClinGen ExAC gnomAD |
|
|
CA8057048 rs778254783 |
68 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746359591 CA8057046 |
72 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA395921850 rs746359591 |
72 | P>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 77 | Y>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 80 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 81 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs980836688 CA281361831 |
86 | V>I | No |
ClinGen TOPMed |
|
|
rs969422442 CA281361822 |
87 | V>L | No |
ClinGen TOPMed |
|
| TCGA novel | 88 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA281361787 rs912559809 |
88 | K>Q | No |
ClinGen gnomAD |
|
|
CA8057022 rs777982018 |
94 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1314598157 CA395921151 |
95 | Y>C | No |
ClinGen gnomAD |
|
|
CA8057020 rs752861266 |
97 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
rs779196597 CA281361742 |
100 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755048837 CA8057018 |
101 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA8057017 COSM3818093 rs753984711 |
101 | R>H | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs755048837 CA395921096 |
101 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 102 | S>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA395921034 rs1200188293 |
105 | M>T | No |
ClinGen gnomAD |
|
|
CA281361654 rs753201275 |
108 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
CA395920995 rs1473245954 |
109 | V>L | No |
ClinGen gnomAD |
|
|
CA395920982 rs1156378312 |
110 | I>T | No |
ClinGen TOPMed |
|
|
CA281361652 rs1032844543 |
113 | R>Q | No |
ClinGen Ensembl |
|
|
rs769347114 CA8057006 |
113 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA395920926 rs1191455290 |
116 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA8057005 rs747506016 |
119 | D>N | No |
ClinGen ExAC gnomAD |
|
|
COSM971478 CA8057003 rs373752479 |
121 | V>I | Variant assessed as Somatic; 0.0 impact. central_nervous_system endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 128 | I>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8057000 rs755278060 |
130 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA395920750 rs1405384807 |
133 | P>A | No |
ClinGen gnomAD |
|
|
rs1177246282 CA395920725 |
135 | G>C | No |
ClinGen gnomAD |
|
|
CA8056999 rs370289246 |
137 | C>G | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1427164346 CA395920699 |
137 | C>Y | No |
ClinGen gnomAD |
|
|
rs781311887 COSM971477 CA8056952 |
139 | R>C | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA8056951 rs757341232 |
139 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA395920082 rs1158115486 |
140 | L>W | No |
ClinGen TOPMed |
|
|
rs953034676 CA281360946 |
141 | V>L | No |
ClinGen TOPMed |
|
|
rs1394013364 CA395920056 |
143 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs758135917 CA8056948 |
144 | I>S | No |
ClinGen ExAC gnomAD |
|
|
rs1300948018 CA395920036 |
146 | V>I | No |
ClinGen TOPMed |
|
|
CA8056947 rs752514939 |
148 | H>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA281360927 rs995013998 |
149 | P>A | No |
ClinGen TOPMed |
|
|
CA8056946 rs778493299 |
149 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs754625474 CA8056945 |
154 | T>A | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 156 | G>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1567757016 CA395919889 |
158 | L>M | No |
ClinGen Ensembl |
|
|
rs765897827 CA8056943 |
160 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 162 | R>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1347930660 CA395919832 |
162 | R>T | No |
ClinGen TOPMed |
|
|
rs760168557 CA8056942 |
165 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA281360715 rs1044968841 |
169 | R>Q | No |
ClinGen TOPMed |
|
|
CA395919708 rs1478076012 |
174 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA281360693 rs61731846 |
175 | W>* | No |
ClinGen Ensembl |
|
|
CA281360697 rs61731847 |
175 | W>L | No |
ClinGen Ensembl |
|
|
rs995190946 CA281360685 |
176 | Q>* | No |
ClinGen Ensembl |
|
|
CA8056920 rs753316244 |
177 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs963725014 CA281360672 |
178 | L>I | No |
ClinGen TOPMed |
|
|
CA395919680 rs1244858630 |
179 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
rs759826964 CA8056918 |
183 | R>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 185 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs776857155 CA8056917 |
189 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs771085637 CA8056916 |
190 | T>R | No |
ClinGen ExAC gnomAD |
|
|
rs150593359 CA281360609 |
191 | A>T | No |
ClinGen ESP TOPMed |
|
|
rs1259542641 CA395919585 |
192 | S>N | No |
ClinGen TOPMed |
|
|
CA395919580 rs1340502168 |
193 | P>S | No |
ClinGen TOPMed |
|
|
rs773241653 CA8056915 |
194 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs773241653 CA8056914 |
194 | L>R | No |
ClinGen ExAC gnomAD |
|
|
rs772184838 CA8056913 |
196 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA281360562 rs1012088845 |
196 | P>L | No |
ClinGen TOPMed |
|
|
rs748061956 CA8056912 |
198 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA8056911 rs778887599 |
200 | V>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 204 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA395919489 rs1313357996 |
205 | D>E | No |
ClinGen TOPMed |
|
| TCGA novel | 205 | D>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1354301019 CA395919484 |
206 | I>T | No |
ClinGen TOPMed |
|
|
rs1260668126 CA395919488 |
206 | I>V | No |
ClinGen Ensembl |
|
|
CA8056894 rs760860184 |
210 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA395919432 rs1194237105 |
213 | V>A | No |
ClinGen gnomAD |
|
| TCGA novel | 213 | V>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA395919435 rs1279710988 |
213 | V>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA281360360 rs767578006 |
214 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767578006 CA8056892 |
214 | V>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8056893 rs773479084 |
214 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs761884339 CA8056891 |
216 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs1193762457 CA395919395 |
219 | V>A | No |
ClinGen gnomAD |
|
|
rs911361305 CA281360351 |
219 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
rs911361305 CA395919397 |
219 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
rs151134332 CA281360343 |
221 | T>A | No |
ClinGen 1000Genomes |
|
|
CA8056890 rs774284749 |
223 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8056889 rs528168629 COSM1378338 |
223 | R>H | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA395919372 rs528168629 |
223 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749069548 CA8056888 |
224 | L>S | No |
ClinGen ExAC gnomAD |
|
|
rs1437625343 CA395919360 |
225 | F>S | No |
ClinGen gnomAD |
|
| rs1347301445 | 225 | F>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA281360319 rs559351790 |
227 | Q>E | No |
ClinGen Ensembl |
|
|
CA395919341 rs1598150377 |
228 | P>S | No |
ClinGen Ensembl |
|
|
CA8056886 rs769769588 |
229 | K>T | No |
ClinGen ExAC |
|
|
rs780998485 CA395919302 |
233 | P>H | No |
ClinGen ExAC gnomAD |
|
|
CA8056884 rs780998485 |
233 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs748916763 CA281360288 |
234 | Y>C | No |
ClinGen Ensembl |
|
|
CA8056881 RCV000900067 rs118126947 |
235 | A>T | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA395919287 rs1427104315 |
236 | I>V | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 237 | S>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8056859 rs374819167 |
240 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA395918589 rs1216332913 |
243 | P>A | No |
ClinGen gnomAD |
|
|
CA8056856 rs767877058 |
246 | H>P | No |
ClinGen ExAC gnomAD |
|
|
rs750806890 CA8056855 |
247 | D>H | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 251 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA395918480 rs1284857214 |
251 | E>D | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 252 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA395918453 rs1448593027 |
254 | L>M | No |
ClinGen TOPMed gnomAD |
|
|
CA395918439 rs1390158105 |
255 | T>I | No |
ClinGen TOPMed |
|
| rs760290777 | 258 | K>missing | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs551631734 CA8056832 |
258 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1327909463 CA395918313 |
261 | E>K | No |
ClinGen TOPMed |
|
| TCGA novel | 261 | E>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs770316581 CA8056830 |
262 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8056829 rs770316581 |
262 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746414525 CA8056828 |
263 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA8056827 rs143575360 |
265 | K>E | No |
ClinGen ESP ExAC TOPMed |
|
|
rs1208224739 CA395918236 |
266 | S>N | No |
ClinGen gnomAD |
|
|
CA8056826 rs757708234 |
267 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs751667313 CA8056825 |
268 | H>Y | No |
ClinGen ExAC |
|
|
rs758363858 CA8056823 |
270 | A>T | No |
ClinGen ExAC |
|
|
CA395918182 rs1488397374 |
271 | G>S | No |
ClinGen gnomAD |
|
|
CA395918159 rs1598146921 |
273 | S>* | No |
ClinGen Ensembl |
|
|
rs201981579 CA8056821 |
276 | K>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8056818 rs145847118 |
280 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1280086197 CA395918082 |
281 | Q>E | No |
ClinGen gnomAD |
|
|
rs1384861610 CA395918073 |
281 | Q>H | No |
ClinGen gnomAD |
|
|
rs568156696 CA8056816 |
289 | T>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA281358661 rs1034577709 |
289 | T>K | No |
ClinGen Ensembl |
|
|
CA8056815 rs772679190 |
290 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772679190 CA395917965 |
290 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771776966 CA8056814 COSM3421035 |
291 | A>V | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1426404310 CA395917930 |
293 | T>K | No |
ClinGen gnomAD |
No associated diseases with Q9H8T0
1 regional properties for Q9H8T0
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Ubiquitin-conjugating enzyme E2 | 74 - 222 | IPR000608 |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| FHF complex | A protein complex that is composed of AKTIP/FTS, FAM160A2/p107FHIP, and one or more members of the Hook family of proteins, HOOK1, HOOK2, and HOOK3. The complex is thought to promote vesicle trafficking and/or fusion, and associates with the homotypic vesicular sorting complex (the HOPS complex). |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| ubiquitin conjugating enzyme activity | Isoenergetic transfer of ubiquitin from one protein to another via the reaction X-ubiquitin + Y -> Y-ubiquitin + X, where both the X-ubiquitin and Y-ubiquitin linkages are thioester bonds between the C-terminal glycine of ubiquitin and a sulfhydryl side group of a cysteine residue. |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| early endosome to late endosome transport | The directed movement of substances, in membrane-bounded vesicles, from the early sorting endosomes to the late sorting endosomes; transport occurs along microtubules and can be experimentally blocked with microtubule-depolymerizing drugs. |
| endosome organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of endosomes. |
| endosome to lysosome transport | The directed movement of substances from endosomes to lysosomes. |
| lysosome organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a lysosome. A lysosome is a cytoplasmic, membrane-bounded organelle that is found in most animal cells and that contains a variety of hydrolases. |
| positive regulation of protein binding | Any process that activates or increases the frequency, rate or extent of protein binding. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| protein localization to perinuclear region of cytoplasm | A process in which a protein is transported to, or maintained in, a location within the perinuclear region of the cytoplasm. |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
18 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5ZJJ5 | AKTIP | AKT-interacting protein | Gallus gallus (Chicken) | PR |
| Q7K4V4 | CG46338 | Protein crossbronx | Drosophila melanogaster (Fruit fly) | PR |
| P25867 | eff | Ubiquitin-conjugating enzyme E2-17 kDa | Drosophila melanogaster (Fruit fly) | PR |
| Q9Y385 | UBE2J1 | Ubiquitin-conjugating enzyme E2 J1 | Homo sapiens (Human) | PR |
| P61086 | UBE2K | Ubiquitin-conjugating enzyme E2 K | Homo sapiens (Human) | PR |
| Q9NPD8 | UBE2T | Ubiquitin-conjugating enzyme E2 T | Homo sapiens (Human) | PR |
| O00762 | UBE2C | Ubiquitin-conjugating enzyme E2 C | Homo sapiens (Human) | PR |
| O14933 | UBE2L6 | Ubiquitin/ISG15-conjugating enzyme E2 L6 | Homo sapiens (Human) | PR |
| Q96LR5 | UBE2E2 | Ubiquitin-conjugating enzyme E2 E2 | Homo sapiens (Human) | PR |
| Q969T4 | UBE2E3 | Ubiquitin-conjugating enzyme E2 E3 | Homo sapiens (Human) | PR |
| Q7Z7E8 | UBE2Q1 | Ubiquitin-conjugating enzyme E2 Q1 | Homo sapiens (Human) | PR |
| A1L167 | UBE2QL1 | Ubiquitin-conjugating enzyme E2Q-like protein 1 | Homo sapiens (Human) | PR |
| Q16763 | UBE2S | Ubiquitin-conjugating enzyme E2 S | Homo sapiens (Human) | EV |
| Q64362 | Aktip | AKT-interacting protein | Mus musculus (Mouse) | PR |
| Q5FVH4 | Aktip | AKT-interacting protein | Rattus norvegicus (Rat) | PR |
| P35129 | let-70 | Ubiquitin-conjugating enzyme E2 2 | Caenorhabditis elegans | PR |
| Q9SLE4 | UBC29 | Ubiquitin-conjugating enzyme E2 29 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C8X7 | UBC31 | Probable ubiquitin-conjugating enzyme E2 31 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNPFWSMSTS | SVRKRSEGEE | KTLTGDVKTS | PPRTAPKKQL | PSIPKNALPI | TKPTSPAPAA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QSTNGTHASY | GPFYLEYSLL | AEFTLVVKQK | LPGVYVQPSY | RSALMWFGVI | FIRHGLYQDG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VFKFTVYIPD | NYPDGDCPRL | VFDIPVFHPL | VDPTSGELDV | KRAFAKWRRN | HNHIWQVLMY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ARRVFYKIDT | ASPLNPEAAV | LYEKDIQLFK | SKVVDSVKVC | TARLFDQPKI | EDPYAISFSP |
| 250 | 260 | 270 | 280 | 290 | |
| WNPSVHDEAR | EKMLTQKKPE | EQHNKSVHVA | GLSWVKPGSV | QPFSKEEKTV | AT |