Q9H082
Gene name |
RAB33B |
Protein name |
Ras-related protein Rab-33B |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:83452 |
EC number |
|
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

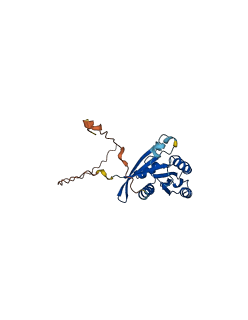
3 structures for Q9H082
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6Y09 | X-ray | 240 A | A/B | 30-218 | PDB |
| 6ZAY | X-ray | 240 A | A/B | 30-218 | PDB |
| AF-Q9H082-F1 | Predicted | AlphaFoldDB |
184 variants for Q9H082
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000488443 rs1085307130 |
17 | G>missing | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV002520202 RCV000323050 CA3083945 rs371561776 RCV002520203 |
43 | N>S | Smith-McCort dysplasia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
VAR_068854 CA143715 rs587776958 RCV000043483 |
46 | K>Q | Smith-McCort dysplasia 2 SMC2 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000735865 rs1561002040 |
63 | E>missing | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA358271927 RCV000488447 rs1085307128 |
71 | R>* | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000488435 rs1085307129 CA358272776 |
122 | F>S | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001352912 RCV002287497 rs1750416123 |
134 | Q>* | Smith-McCort dysplasia 2 Skeletal dysplasia [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001861226 RCV000383050 CA3084015 rs200448316 |
139 | N>K | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs886044716 CA10602368 VAR_068855 RCV000043484 |
148 | N>K | Smith-McCort dysplasia 2 SMC2; strongly inhibits protein expression and may disrupt the Golgi apparatus structure [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000488449 rs1085307131 CA358273047 |
164 | Q>* | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001573007 RCV001148162 rs140381459 CA3084027 |
177 | T>M | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA3084032 rs747031994 RCV000343754 |
185 | N>D | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV002061241 RCV000389054 CA3084046 rs139823051 |
226 | T>M | Smith-McCort dysplasia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs1561001821 CA358271437 |
3 | E>G | No |
ClinGen Ensembl |
|
|
CA358271435 rs1561001821 |
3 | E>V | No |
ClinGen Ensembl |
|
|
rs757940962 CA3083922 |
4 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA358271472 rs1313626293 CA358271471 |
5 | M>I | No |
ClinGen TOPMed |
|
|
rs886251366 CA358271465 |
5 | M>K | No |
ClinGen TOPMed gnomAD |
|
|
CA106678346 rs886251366 |
5 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1579171628 CA358271497 |
7 | S>A | No |
ClinGen Ensembl |
|
|
CA3083923 rs377532895 |
7 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 8 | S>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1242581833 CA358271515 |
8 | S>L | No |
ClinGen gnomAD |
|
|
rs1165656260 CA358271520 |
9 | L>F | No |
ClinGen gnomAD |
|
|
CA3083925 rs755362234 |
9 | L>H | No |
ClinGen ExAC gnomAD |
|
|
rs368228699 CA3083928 |
10 | E>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3083927 rs748593700 |
10 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA358271532 rs748593700 |
10 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs974858262 CA106678361 |
13 | F>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1449868596 CA358271582 |
16 | S>C | No |
ClinGen gnomAD |
|
|
rs1316627107 CA358271585 |
16 | S>I | No |
ClinGen gnomAD |
|
|
CA106678375 rs921925813 |
17 | G>E | No |
ClinGen TOPMed |
|
|
CA358271595 rs1318546831 |
18 | A>E | No |
ClinGen gnomAD |
|
|
CA3083931 rs770964994 |
18 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs759734964 CA3083933 |
22 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs776574187 CA3083935 |
24 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA358271634 rs1579171725 |
25 | F>V | No |
ClinGen Ensembl |
|
|
rs1579171732 CA358271643 |
26 | L>S | No |
ClinGen Ensembl |
|
|
rs765290544 CA3083937 |
27 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA358271654 rs1410588428 |
28 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1401671327 CA358271657 |
28 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1410588428 CA358271653 |
28 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1176282934 CA358271658 |
29 | A>T | No |
ClinGen gnomAD |
|
|
rs762497408 CA3083939 |
30 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1317573710 CA358271674 |
31 | S>F | No |
ClinGen TOPMed |
|
|
CA358271677 rs1338119168 |
32 | R>C | No |
ClinGen gnomAD |
|
|
rs1293950567 CA358271678 |
32 | R>H | No |
ClinGen TOPMed |
|
|
CA3083941 rs766581102 |
33 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs367812678 CA3083942 |
34 | F>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 41 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1224161471 CA358271745 |
42 | S>A | No |
ClinGen gnomAD |
|
|
CA106678519 rs980175514 |
43 | N>H | No |
ClinGen TOPMed |
|
|
rs371561776 CA358271751 |
43 | N>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1418167506 CA358271770 |
46 | K>R | No |
ClinGen TOPMed |
|
|
CA3083949 rs753943143 |
49 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA358271806 rs1202632463 |
52 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1202632463 CA358271807 |
52 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs746037476 CA3083950 |
54 | C>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3083952 rs776521254 |
55 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs772457148 CA3083951 |
55 | A>S | No |
ClinGen ExAC |
|
|
rs1481705399 CA358271834 |
56 | G>D | No |
ClinGen gnomAD |
|
|
CA3083954 rs149649513 |
56 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA358271838 rs1402270892 |
57 | R>C | No |
ClinGen gnomAD |
|
|
CA358271840 rs773200487 |
57 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA3083955 rs773200487 |
57 | R>P | No |
ClinGen ExAC gnomAD |
|
|
CA358271845 rs1457451598 |
58 | F>S | No |
ClinGen TOPMed |
|
|
CA358271852 rs1274061972 |
59 | P>R | No |
ClinGen TOPMed |
|
|
rs762852071 CA3083956 |
59 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA358271854 rs1355924118 |
60 | D>Y | No |
ClinGen gnomAD |
|
|
CA106678676 rs867952323 |
61 | R>H | No |
ClinGen Ensembl |
|
|
CA358271871 rs1410142464 |
62 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
CA358271873 rs1262766150 |
63 | E>K | No |
ClinGen gnomAD |
|
|
rs755124749 CA3083958 |
64 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA3083959 rs759147084 |
67 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA358271909 rs1579171940 |
68 | V>G | No |
ClinGen Ensembl |
|
|
CA358271916 rs1212883458 |
69 | D>E | No |
ClinGen gnomAD |
|
|
CA358271925 rs752394542 |
70 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA358271926 rs1085307128 |
71 | R>G | No |
ClinGen gnomAD |
|
| TCGA novel | 71 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA106678698 rs1035382343 |
73 | R>G | No |
ClinGen TOPMed |
|
|
CA358271942 rs1222118960 |
73 | R>L | No |
ClinGen gnomAD |
|
|
rs1267179477 CA358271948 |
74 | A>V | No |
ClinGen gnomAD |
|
|
CA3083963 rs764433053 |
75 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA358271949 rs764433053 |
75 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3083964 rs202128260 |
77 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA358271969 rs1394696036 |
78 | D>Y | No |
ClinGen TOPMed |
|
|
CA106678742 rs886674153 |
79 | G>E | No |
ClinGen TOPMed gnomAD |
|
|
CA358271980 rs886674153 |
79 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
CA358271983 COSM3380814 rs1421096342 |
80 | E>K | pancreas [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA106678746 rs911276815 |
81 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs757709966 CA358271991 |
81 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs757709966 CA3083965 |
81 | R>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 82 | I>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3083966 rs779011534 |
82 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1335371166 CA358272508 |
84 | I>V | No |
ClinGen TOPMed |
|
|
CA106694727 rs144431471 |
85 | Q>R | No |
ClinGen ESP |
|
|
rs1464902557 CA358272538 |
88 | D>V | No |
ClinGen TOPMed gnomAD |
|
|
CA358272536 rs1285611461 |
88 | D>Y | No |
ClinGen gnomAD |
|
|
CA3083993 rs778683662 |
89 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA358272547 rs1240788335 |
90 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 91 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 91 | G>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1187861686 CA358272576 |
94 | R>* | No |
ClinGen gnomAD |
|
|
rs1389587674 CA358272577 |
94 | R>Q | No |
ClinGen TOPMed |
|
|
CA358272630 rs1561008272 |
101 | Q>L | No |
ClinGen Ensembl |
|
|
rs774986521 CA3083997 |
104 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs554278184 CA3083999 |
108 | H>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs776396935 CA3084000 |
109 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1241162788 CA358272690 |
110 | V>I | No |
ClinGen TOPMed |
|
|
CA358272707 rs1396804009 |
112 | F>S | No |
ClinGen gnomAD |
|
|
rs150536664 CA3084002 |
113 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA106694824 rs901826209 |
114 | Y>N | No |
ClinGen Ensembl |
|
|
rs1289459041 CA358272724 |
115 | D>Y | No |
ClinGen gnomAD |
|
|
rs1579183594 CA358272758 |
119 | M>I | No |
ClinGen Ensembl |
|
|
CA358272755 rs1323432245 |
119 | M>L | No |
ClinGen gnomAD |
|
|
CA3084004 rs763571730 |
119 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775417496 CA106694847 |
120 | A>S | No |
ClinGen Ensembl |
|
|
CA3084005 rs766496342 |
121 | S>I | No |
ClinGen ExAC gnomAD |
|
|
rs751716862 CA3084006 |
123 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA3084007 rs139890586 |
127 | S>F | No |
ClinGen ESP ExAC gnomAD |
|
| TCGA novel | 127 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs200395089 CA3084008 |
129 | I>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV002741704 CA358272837 rs1295900136 |
131 | E>* | No |
ClinGen ClinVar dbSNP gnomAD |
|
|
CA3084009 rs753002792 |
132 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
CA3084010 rs756996980 |
133 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA106694984 rs997966212 |
135 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA3084012 rs372938614 |
136 | L>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA3084014 rs779490913 |
139 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763106736 CA3084016 |
141 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA106695007 rs369719131 |
142 | P>L | No |
ClinGen Ensembl |
|
|
CA358272915 rs1345316876 |
143 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs776147025 CA3084017 |
143 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1313401877 CA358272932 |
146 | V>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1313401877 CA358272933 |
146 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA358272945 rs1233351667 |
148 | N>D | No |
ClinGen gnomAD |
|
|
CA106695030 rs201596716 |
148 | N>S | No |
ClinGen gnomAD |
|
|
CA358272958 rs1206938913 |
150 | C>R | No |
ClinGen TOPMed gnomAD |
|
|
rs773712674 CA3084020 |
151 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA3084022 CA3084021 rs763510197 |
154 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs997046063 CA106695065 |
155 | A>V | No |
ClinGen TOPMed |
|
|
CA106695102 rs374903402 |
156 | I>K | No |
ClinGen ESP |
|
|
rs374903402 CA106695112 |
156 | I>T | No |
ClinGen ESP |
|
|
rs143365276 CA106695080 |
156 | I>V | No |
ClinGen ESP |
|
|
CA358273005 rs1426612032 |
157 | Q>R | No |
ClinGen TOPMed |
|
|
rs1431359197 CA358273011 |
158 | V>L | No |
ClinGen gnomAD |
|
|
rs369144894 CA3084024 |
159 | P>R | No |
ClinGen ESP ExAC gnomAD |
|
| TCGA novel | 160 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3084025 rs767704176 |
160 | T>R | No |
ClinGen ExAC gnomAD |
|
|
CA106695134 rs111577712 |
162 | L>S | No |
ClinGen Ensembl |
|
|
CA358273080 rs1365292099 |
168 | D>G | No |
ClinGen gnomAD |
|
|
rs1579183742 CA358273076 RCV000998298 |
168 | D>N | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1482631626 CA358273099 |
171 | S>C | No |
ClinGen TOPMed |
|
|
CA358273114 rs1403526632 |
172 | M>I | No |
ClinGen gnomAD |
|
|
CA106695140 rs993911491 |
178 | S>F | No |
ClinGen Ensembl |
|
|
CA358273173 rs1288530182 |
181 | N>T | No |
ClinGen TOPMed |
|
|
CA3084030 rs758237954 |
182 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA358273176 rs758237954 |
182 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3084031 rs779934522 |
183 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs754430101 CA3084033 |
185 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs769605100 CA3084037 |
187 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3084038 rs749651332 |
190 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771464960 CA3084039 |
193 | M>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3084040 rs774996373 |
196 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1426792991 CA358273279 |
197 | H>Y | No |
ClinGen TOPMed |
|
|
CA3084041 rs760149110 |
202 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1579183832 CA358273335 |
204 | P>L | No |
ClinGen Ensembl |
|
|
CA358273351 rs1165851890 |
206 | M>I | No |
ClinGen gnomAD |
|
|
rs1350898847 CA358273361 |
208 | S>N | No |
ClinGen gnomAD |
|
|
rs1453636456 CA358273376 |
210 | P>S | No |
ClinGen gnomAD |
|
|
CA358273383 rs1408459156 |
211 | P>H | No |
ClinGen TOPMed gnomAD |
|
|
CA358273385 rs1408459156 |
211 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1408459156 CA358273384 |
211 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
CA358273380 rs1359202074 |
211 | P>S | No |
ClinGen gnomAD |
|
|
rs1436649466 CA358273387 |
212 | D>H | No |
ClinGen gnomAD |
|
|
CA358273399 rs1201049287 |
213 | N>S | No |
ClinGen TOPMed |
|
|
CA3084043 rs775643958 |
216 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs1489736435 CA358273422 |
217 | L>V | No |
ClinGen TOPMed |
|
|
CA358273438 rs1156531467 |
219 | P>L | No |
ClinGen gnomAD |
|
|
rs201952341 CA3084044 |
219 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA358273458 rs1318239462 |
222 | K>M | No |
ClinGen TOPMed |
|
|
CA3084045 rs764278121 |
224 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1291716060 CA358273473 |
225 | M>V | No |
ClinGen gnomAD |
|
|
rs1189315237 CA358273500 |
228 | W>C | No |
ClinGen gnomAD |
|
|
rs751390982 CA3084049 |
228 | W>R | No |
ClinGen ExAC gnomAD |
1 associated diseases with Q9H082
[MIM: 615222]: Smith-McCort dysplasia 2 (SMC2)
A rare autosomal recessive osteochondrodysplasia with skeletal features identical to those of Dyggve-Melchior-Clausen syndrome, but with normal intelligence and no microcephaly. It is characterized by short limbs and trunk with barrel-shaped chest. The radiographic phenotype includes platyspondyly, generalized abnormalities of the epiphyses and metaphyses, and a distinctive lacy appearance of the iliac crest. {ECO:0000269|PubMed:22652534, ECO:0000269|PubMed:23042644}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A rare autosomal recessive osteochondrodysplasia with skeletal features identical to those of Dyggve-Melchior-Clausen syndrome, but with normal intelligence and no microcephaly. It is characterized by short limbs and trunk with barrel-shaped chest. The radiographic phenotype includes platyspondyly, generalized abnormalities of the epiphyses and metaphyses, and a distinctive lacy appearance of the iliac crest. {ECO:0000269|PubMed:22652534, ECO:0000269|PubMed:23042644}. Note=The disease is caused by variants affecting the gene represented in this entry.
1 regional properties for Q9H082
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 33 - 181 | IPR005225 |
Functions
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| Golgi lumen | The volume enclosed by the membranes of any cisterna or subcompartment of the Golgi apparatus, including the cis- and trans-Golgi networks. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome assembly | The formation of a double membrane-bounded structure, the autophagosome, that occurs when a specialized membrane sac, called the isolation membrane, starts to enclose a portion of the cytoplasm. |
| intra-Golgi vesicle-mediated transport | The directed movement of substances within the Golgi, mediated by small transport vesicles. These either fuse with the cis-Golgi or with each other to form the membrane stacks known as the cis-Golgi reticulum (network). |
| negative regulation of constitutive secretory pathway | Any process that stops, prevents or reduces the frequency, rate or extent of constitutive secretory pathway. |
| protein localization to Golgi apparatus | A process in which a protein is transported to, or maintained in, a location within the Golgi apparatus. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| Rab protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Rab family of proteins switching to a GTP-bound active state. |
| regulation of exocytosis | Any process that modulates the frequency, rate or extent of exocytosis. |
| regulation of Golgi organization | Any process that modulates the frequency, rate or extent of Golgi organization. |
| regulation of retrograde vesicle-mediated transport, Golgi to ER | Any process that modulates the frequency, rate or extent of retrograde vesicle-mediated transport, Golgi to ER. |
| skeletal system morphogenesis | The process in which the anatomical structures of the skeleton are generated and organized. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3ZC27 | RAB19 | Ras-related protein Rab-19 | Bos taurus (Bovine) | PR |
| Q5ZHV1 | RAB33B | Ras-related protein Rab-33B | Gallus gallus (Chicken) | PR |
| A4D1S5 | RAB19 | Ras-related protein Rab-19 | Homo sapiens (Human) | PR |
| Q96AX2 | RAB37 | Ras-related protein Rab-37 | Homo sapiens (Human) | PR |
| P35294 | Rab19 | Ras-related protein Rab-19 | Mus musculus (Mouse) | PR |
| O35963 | Rab33b | Ras-related protein Rab-33B | Mus musculus (Mouse) | PR |
| Q5M7U5 | Rab19 | Ras-related protein Rab-19 | Rattus norvegicus (Rat) | PR |
| P25766 | RGP1 | Ras-related protein RGP1 | Oryza sativa subsp japonica (Rice) | PR |
| Q20365 | rab-33 | Ras-related protein Rab-33 | Caenorhabditis elegans | PR |
| Q9LNK1 | RABA3 | Ras-related protein RABA3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O04486 | RABA2A | Ras-related protein RABA2a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LNW1 | RABA2B | Ras-related protein RABA2b | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LH50 | RABA4D | Ras-related protein RABA4d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FE79 | RABA4C | Ras-related protein RABA4c | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJZ5 | RABA4E | Putative Ras-related protein RABA4e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAEEMESSLE | ASFSSSGAVS | GASGFLPPAR | SRIFKIIVIG | DSNVGKTCLT | YRFCAGRFPD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RTEATIGVDF | RERAVEIDGE | RIKIQLWDTA | GQERFRKSMV | QHYYRNVHAV | VFVYDMTNMA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SFHSLPSWIE | ECKQHLLAND | IPRILVGNKC | DLRSAIQVPT | DLAQKFADTH | SMPLFETSAK |
| 190 | 200 | 210 | 220 | ||
| NPNDNDHVEA | IFMTLAHKLK | SHKPLMLSQP | PDNGIILKPE | PKPAMTCWC |