Q9FNE1
Gene name |
CRK42 (At5g40380, MPO12.90) |
Protein name |
Cysteine-rich receptor-like protein kinase 42 |
Names |
Cysteine-rich RLK42 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G40380 |
EC number |
|
Protein Class |
|
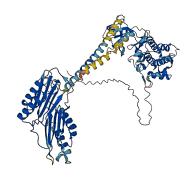
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
457-464 (Activation loop from InterPro)
Target domain |
315-604 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
475-481 (Activation loop from InterPro)
Target domain |
315-604 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9FNE1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9FNE1-F1 | Predicted | AlphaFoldDB |
101 variants for Q9FNE1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH07278852 | 2 | R>H | No | 1000Genomes | |
| ENSVATH07278853 | 7 | T>S | No | 1000Genomes | |
| ENSVATH12277240 | 13 | V>I | No | 1000Genomes | |
| ENSVATH07278854 | 17 | Y>H | No | 1000Genomes | |
| tmp_5_16152173_CCTT_GCTT,C | 18 | S>C | No | 1000Genomes | |
| ENSVATH12277241 | 21 | F>I | No | 1000Genomes | |
| tmp_5_16152205_T_G | 29 | S>A | No | 1000Genomes | |
| tmp_5_16152206_C_A | 29 | S>Y | No | 1000Genomes | |
| ENSVATH12277242 | 45 | S>I | No | 1000Genomes | |
| ENSVATH07278857 | 56 | T>N | No | 1000Genomes | |
| ENSVATH03300830 | 75 | T>I | No | 1000Genomes | |
| ENSVATH03300830 | 75 | T>N | No | 1000Genomes | |
| ENSVATH07278858 | 76 | E>K | No | 1000Genomes | |
| ENSVATH03300831 | 81 | T>S | No | 1000Genomes | |
| ENSVATH14555656 | 97 | S>F | No | 1000Genomes | |
| tmp_5_16152419_A_G | 100 | Q>R | No | 1000Genomes | |
| ENSVATH03300832 | 110 | I>L | No | 1000Genomes | |
| tmp_5_16152506_G_A | 129 | R>H | No | 1000Genomes | |
| tmp_5_16152520_G_C | 134 | E>Q | No | 1000Genomes | |
| ENSVATH00690663 | 143 | A>T | No | 1000Genomes | |
| ENSVATH00690665 | 153 | T>I | No | 1000Genomes | |
| ENSVATH03300835 | 156 | D>N | No | 1000Genomes | |
| ENSVATH03300836 | 157 | P>L | No | 1000Genomes | |
| tmp_5_16152593_G_A | 158 | R>Q | No | 1000Genomes | |
| ENSVATH12277244 | 168 | A>V | No | 1000Genomes | |
| ENSVATH03300837 | 170 | V>L | No | 1000Genomes | |
| ENSVATH14555658 | 173 | R>K | No | 1000Genomes | |
| ENSVATH00690666 | 183 | N>S | No | 1000Genomes | |
| tmp_5_16152673_G_A | 185 | V>I | No | 1000Genomes | |
| ENSVATH12277277 | 205 | E>D | No | 1000Genomes | |
| tmp_5_16152733_G_A | 205 | E>K | No | 1000Genomes | |
| ENSVATH12277278 | 222 | A>V | No | 1000Genomes | |
| tmp_5_16152794_C_G | 225 | T>R | No | 1000Genomes | |
| ENSVATH07278863 | 230 | R>K | No | 1000Genomes | |
| ENSVATH14555659 | 232 | S>P | No | 1000Genomes | |
| ENSVATH12277279 | 243 | H>R | No | 1000Genomes | |
| tmp_5_16152857_A_G | 246 | H>R | No | 1000Genomes | |
| ENSVATH00690668 | 248 | L>H | No | 1000Genomes | |
| ENSVATH07278882 | 254 | I>V | No | 1000Genomes | |
| tmp_5_16153182_C_T | 261 | T>M | No | 1000Genomes | |
| tmp_5_16153205_C_G | 269 | L>V | No | 1000Genomes | |
| ENSVATH03300860 | 270 | L>S | No | 1000Genomes | |
| ENSVATH00690669 | 274 | V>A | No | 1000Genomes | |
| ENSVATH03300861 | 277 | T>I | No | 1000Genomes | |
| ENSVATH03300863 | 280 | S>L | No | 1000Genomes | |
| tmp_5_16153494_A_G | 285 | E>G | No | 1000Genomes | |
| ENSVATH07278890 | 288 | N>S | No | 1000Genomes | |
| ENSVATH00690673 | 294 | R>K | No | 1000Genomes | |
| ENSVATH03300882 | 295 | K>N | No | 1000Genomes | |
| tmp_5_16153541_A_G | 301 | T>A | No | 1000Genomes | |
| ENSVATH07278891 | 304 | K>R | No | 1000Genomes | |
| ENSVATH14555665 | 311 | A>G | No | 1000Genomes | |
| ENSVATH12277349 | 312 | T>S | No | 1000Genomes | |
| ENSVATH07278892 | 313 | D>Y | No | 1000Genomes | |
| ENSVATH03300886 | 317 | H>P | No | 1000Genomes | |
| ENSVATH00690674 | 318 | K>T | No | 1000Genomes | |
| ENSVATH03300888 | 320 | M>I | No | 1000Genomes | |
| ENSVATH03300887 | 320 | M>T | No | 1000Genomes | |
| ENSVATH00690675 | 336 | N>T | No | 1000Genomes | |
| ENSVATH03300890 | 337 | G>C | No | 1000Genomes | |
| ENSVATH00690676 | 338 | K>N | No | 1000Genomes | |
| ENSVATH03300891 | 351 | D>E | No | 1000Genomes | |
| ENSVATH07278893 | 356 | F>V | No | 1000Genomes | |
| ENSVATH07278896 | 407 | K>N | No | 1000Genomes | |
| ENSVATH00690678 | 412 | S>R | No | 1000Genomes | |
| tmp_5_16154182_C_T | 432 | S>F | No | 1000Genomes | |
| tmp_5_16154215_C_G | 443 | T>S | No | 1000Genomes | |
| tmp_5_16154251_A_T | 455 | K>M | No | 1000Genomes | |
| tmp_5_16154305_G_C | 473 | S>T | No | 1000Genomes | |
| ENSVATH03300906 | 490 | R>T | No | 1000Genomes | |
| tmp_5_16154485_C_T | 497 | A>V | No | 1000Genomes | |
| ENSVATH07278903 | 510 | A>V | No | 1000Genomes | |
| ENSVATH12277437 | 513 | T>I | No | 1000Genomes | |
| ENSVATH12277438 | 539 | V>I | No | 1000Genomes | |
| ENSVATH03300909 | 540 | E>K | No | 1000Genomes | |
| tmp_5_16154703_G_A | 541 | A>T | No | 1000Genomes | |
| ENSVATH07278907 | 543 | D>E | No | 1000Genomes | |
| ENSVATH07278908 | 544 | P>T | No | 1000Genomes | |
| ENSVATH03300910 | 545 | C>F | No | 1000Genomes | |
| ENSVATH14555669 | 548 | D>H | No | 1000Genomes | |
| ENSVATH03300911 | 550 | F>L | No | 1000Genomes | |
| ENSVATH14555670 | 560 | A>V | No | 1000Genomes | |
| ENSVATH07278910 | 564 | L>F | No | 1000Genomes | |
| ENSVATH03300912 | 571 | T>A | No | 1000Genomes | |
| tmp_5_16154799_G_C | 573 | A>P | No | 1000Genomes | |
| tmp_5_16154805_C_T | 575 | P>S | No | 1000Genomes | |
| tmp_5_16154809_C_T | 576 | S>L | No | 1000Genomes | |
| ENSVATH12277439 | 581 | M>I | No | 1000Genomes | |
| tmp_5_16154839_G_A | 586 | R>H | No | 1000Genomes | |
| ENSVATH12277440 | 591 | R>S | No | 1000Genomes | |
| ENSVATH07278912 | 610 | T>N | No | 1000Genomes | |
| ENSVATH03300917 | 619 | I>L | No | 1000Genomes | |
| ENSVATH14555672 | 627 | T>M | No | 1000Genomes | |
| ENSVATH07278914 | 629 | F>L | No | 1000Genomes | |
| tmp_5_16154988_G_T | 636 | D>Y | No | 1000Genomes | |
| tmp_5_16154992_A_C | 637 | Q>P | No | 1000Genomes | |
| ENSVATH12277441 | 641 | T>S | No | 1000Genomes | |
| ENSVATH12277442 | 647 | T>N | No | 1000Genomes | |
| ENSVATH03300919 | 650 | T>S | No | 1000Genomes | |
| ENSVATH12277443 | 651 | I>M | No | 1000Genomes | |
| ENSVATH03300920 | 652 | I>Y | No | 1000Genomes |
No associated diseases with Q9FNE1
5 regional properties for Q9FNE1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 315 - 604 | IPR000719 |
| domain | Gnk2-homologous domain | 35 - 135 | IPR002902-1 |
| domain | Gnk2-homologous domain | 137 - 236 | IPR002902-2 |
| active_site | Serine/threonine-protein kinase, active site | 436 - 448 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 321 - 343 | IPR017441 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8AXY6 | MUSK | Muscle, skeletal receptor tyrosine protein kinase | Gallus gallus (Chicken) | SS |
| Q24488 | Ror | Tyrosine-protein kinase transmembrane receptor Ror | Drosophila melanogaster (Fruit fly) | SS |
| Q9V6K3 | Nrk | Tyrosine-protein kinase transmembrane receptor Ror2 | Drosophila melanogaster (Fruit fly) | SS |
| P30530 | AXL | Tyrosine-protein kinase receptor UFO | Homo sapiens (Human) | PR |
| Q01973 | ROR1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Homo sapiens (Human) | PR |
| Q01974 | ROR2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Homo sapiens (Human) | EV |
| P18910 | Npr1 | Atrial natriuretic peptide receptor 1 | Rattus norvegicus (Rat) | PR |
| P51842 | Gucy2f | Retinal guanylyl cyclase 2 | Rattus norvegicus (Rat) | PR |
| P16067 | Npr2 | Atrial natriuretic peptide receptor 2 | Rattus norvegicus (Rat) | PR |
| O13146 | epha3 | Ephrin type-A receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRCLTKTRSF | HYVIIFYSFF | FLPFLSSSSD | DQRTTVSGLF | CGGRSKSSAD | PNYIPTFVED |
| 70 | 80 | 90 | 100 | 110 | 120 |
| MHSLSLKLTT | RRFATESLNS | TTSIYALIQC | HDDLSPSDCQ | LCYAIARTRI | PRCLPSSSAR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IFLDGCFLRY | ETYEFYDESV | SDASDSFSCS | NDTVLDPRFG | FQVSETAARV | AVRKGGFGVA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GENGVHALAQ | CWESLGKEDC | RVCLEKAVKE | VKRCVSRREG | RAMNTGCYLR | YSDHKFYNGD |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GHHKFHVLFN | KGVIVAIVLT | TSAFVMLILL | ATYVIMTKVS | KTKQEKRNLG | LVSRKFNNSK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TKFKYETLEK | ATDYFSHKKM | LGQGGNGTVF | LGILPNGKNV | AVKRLVFNTR | DWVEEFFNEV |
| 370 | 380 | 390 | 400 | 410 | 420 |
| NLISGIQHKN | LVKLLGCSIE | GPESLLVYEY | VPNKSLDQFL | FDESQSKVLN | WSQRLNIILG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TAEGLAYLHG | GSPVRIIHRD | IKTSNVLLDD | QLNPKIADFG | LARCFGLDKT | HLSTGIAGTL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GYMAPEYVVR | GQLTEKADVY | SFGVLVLEIA | CGTRINAFVP | ETGHLLQRVW | NLYTLNRLVE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ALDPCLKDEF | LQVQGSEAEA | CKVLRVGLLC | TQASPSLRPS | MEEVIRMLTE | RDYPIPSPTS |
| 610 | 620 | 630 | 640 | 650 | |
| PPFLRVSSLT | TDLEGSSTIS | HSTNSTTTFN | TMVKTDQASY | TSSESSTTRT | I |