Q9FN93
Gene name |
At5g59680 (MTH12.14) |
Protein name |
Probable LRR receptor-like serine/threonine-protein kinase At5g59680 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G59680 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
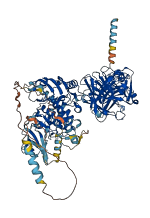
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9FN93
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9FN93-F1 | Predicted | AlphaFoldDB |
83 variants for Q9FN93
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH08581591 | 13 | R>G | No | 1000Genomes | |
| tmp_5_24046835_T_C | 15 | L>S | No | 1000Genomes | |
| ENSVATH14641202 | 47 | P>L | No | 1000Genomes | |
| tmp_5_24047050_G_C | 50 | G>R | No | 1000Genomes | |
| ENSVATH08581607 | 65 | I>N | No | 1000Genomes | |
| ENSVATH08581607 | 65 | I>T | No | 1000Genomes | |
| ENSVATH09557790 | 68 | I>L | No | 1000Genomes | |
| tmp_5_24047127_C_A | 75 | D>E | No | 1000Genomes | |
| ENSVATH00743302 | 119 | D>N | No | 1000Genomes | |
| tmp_5_24047306_C_T | 135 | T>M | No | 1000Genomes | |
| ENSVATH12863665 | 151 | H>Y | No | 1000Genomes | |
| ENSVATH03454210 | 157 | K>N | No | 1000Genomes | |
| ENSVATH08581621 | 177 | V>I | No | 1000Genomes | |
| ENSVATH07451383 | 182 | S>N | No | 1000Genomes | |
| tmp_5_24047496_A_C | 198 | E>D | No | 1000Genomes | |
| ENSVATH03454215 | 228 | Q>H | No | 1000Genomes | |
| ENSVATH03454216 | 230 | N>Y | No | 1000Genomes | |
| ENSVATH12863761 | 237 | N>I | No | 1000Genomes | |
| ENSVATH12863762 | 238 | S>F | No | 1000Genomes | |
| tmp_5_24047932_A_G | 249 | T>A | No | 1000Genomes | |
| ENSVATH00743309 | 260 | P>Q | No | 1000Genomes | |
| ENSVATH00743310 | 273 | Q>K | No | 1000Genomes | |
| ENSVATH03454218 | 315 | S>C | No | 1000Genomes | |
| ENSVATH03454219 | 315 | S>T | No | 1000Genomes | |
| ENSVATH03454220 | 327 | G>E | No | 1000Genomes | |
| tmp_5_24048223_C_T | 346 | L>F | No | 1000Genomes | |
| tmp_5_24048241_T_A | 352 | Y>N | No | 1000Genomes | |
| ENSVATH03454233 | 372 | N>S | No | 1000Genomes | |
| ENSVATH03454237 | 376 | T>S | No | 1000Genomes | |
| ENSVATH12863763 | 380 | S>C | No | 1000Genomes | |
| ENSVATH03454239 | 386 | S>G | No | 1000Genomes | |
| tmp_5_24048522_T_G | 399 | L>V | No | 1000Genomes | |
| ENSVATH12863815 | 417 | L>F | No | 1000Genomes | |
| ENSVATH00743314 | 427 | T>A | No | 1000Genomes | |
| ENSVATH08581687 | 435 | T>S | No | 1000Genomes | |
| tmp_5_24048726_A_G | 438 | K>E | No | 1000Genomes | |
| ENSVATH12863820 | 479 | Q>E | No | 1000Genomes | |
| ENSVATH03454260 | 479 | Q>H | No | 1000Genomes | |
| tmp_5_24049055_G_C | 484 | E>Q | No | 1000Genomes | |
| ENSVATH03454262 | 486 | L>V | No | 1000Genomes | |
| ENSVATH07451407 | 494 | I>T | No | 1000Genomes | |
| ENSVATH07451409 | 508 | P>L | No | 1000Genomes | |
| ENSVATH12863823 | 510 | T>A | No | 1000Genomes | |
| ENSVATH12863824 | 510 | T>I | No | 1000Genomes | |
| ENSVATH12863824 | 510 | T>K | No | 1000Genomes | |
| ENSVATH07451410 | 522 | I>V | No | 1000Genomes | |
| ENSVATH12863845 | 524 | V>L | No | 1000Genomes | |
| ENSVATH03454275 | 525 | L>V | No | 1000Genomes | |
| ENSVATH08581713 | 528 | V>I | No | 1000Genomes | |
| ENSVATH08581713 | 528 | V>L | No | 1000Genomes | |
| ENSVATH07451412 | 530 | F>V | No | 1000Genomes | |
| ENSVATH09557865 | 538 | A>T | No | 1000Genomes | |
| ENSVATH00743315 | 543 | L>H | No | 1000Genomes | |
| ENSVATH03454279 | 556 | A>V | No | 1000Genomes | |
| ENSVATH03454280 | 569 | K>R | No | 1000Genomes | |
| ENSVATH03454281 | 573 | S>L | No | 1000Genomes | |
| ENSVATH03454282 | 576 | T>M | No | 1000Genomes | |
| ENSVATH03454285 | 581 | N>H | No | 1000Genomes | |
| ENSVATH03454287 | 583 | G>E | No | 1000Genomes | |
| ENSVATH03454315 | 645 | E>D | No | 1000Genomes | |
| ENSVATH08581725 | 647 | D>E | No | 1000Genomes | |
| ENSVATH07451422 | 648 | H>D | No | 1000Genomes | |
| ENSVATH12863916 | 669 | G>R | No | 1000Genomes | |
| ENSVATH00743322 | 671 | K>E | No | 1000Genomes | |
| ENSVATH00743322 | 671 | K>Q | No | 1000Genomes | |
| ENSVATH00743323 | 684 | A>V | No | 1000Genomes | |
| ENSVATH03454344 | 736 | S>T | No | 1000Genomes | |
| ENSVATH03454350 | 755 | H>Q | No | 1000Genomes | |
| tmp_5_24050409_C_G | 758 | R>G | No | 1000Genomes | |
| ENSVATH03454352 | 760 | S>G | No | 1000Genomes | |
| ENSVATH03454357 | 777 | T>S | No | 1000Genomes | |
| tmp_5_24050493_C_G | 786 | R>G | No | 1000Genomes | |
| tmp_5_24050527_C_A | 797 | S>Y | No | 1000Genomes | |
| tmp_5_24050550_G_A | 805 | A>T | No | 1000Genomes | |
| ENSVATH00743328 | 810 | L>P | No | 1000Genomes | |
| ENSVATH00743332 | 838 | R>K | No | 1000Genomes | |
| tmp_5_24050703_G_C | 856 | E>Q | No | 1000Genomes | |
| tmp_5_24050728_G_A | 864 | R>Q | No | 1000Genomes | |
| tmp_5_24050730_G_A | 865 | G>S | No | 1000Genomes | |
| tmp_5_24050736_G_T | 867 | D>Y | No | 1000Genomes | |
| ENSVATH03454370 | 868 | T>S | No | 1000Genomes | |
| tmp_5_24050746_G_A | 870 | S>N | No | 1000Genomes | |
| ENSVATH14641203 | 883 | I>F | No | 1000Genomes |
No associated diseases with Q9FN93
5 regional properties for Q9FN93
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 580 - 853 | IPR000719 |
| repeat | Leucine-rich repeat | 410 - 470 | IPR001611 |
| active_site | Serine/threonine-protein kinase, active site | 701 - 713 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 586 - 608 | IPR017441 |
| domain | Malectin-like domain | 32 - 354 | IPR024788 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
35 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q91044 | NTRK3 | NT-3 growth factor receptor | Gallus gallus (Chicken) | SS |
| Q91987 | NTRK2 | BDNF/NT-3 growth factors receptor | Gallus gallus (Chicken) | SS |
| Q91009 | NTRK1 | High affinity nerve growth factor receptor | Gallus gallus (Chicken) | SS |
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| P20594 | NPR2 | Atrial natriuretic peptide receptor 2 | Homo sapiens (Human) | PR |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P24786 | NTRK3 | NT-3 growth factor receptor | Sus scrofa (Pig) | SS |
| P35739 | Ntrk1 | High affinity nerve growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q03351 | Ntrk3 | NT-3 growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q63604 | Ntrk2 | BDNF/NT-3 growth factors receptor | Rattus norvegicus (Rat) | SS |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGG3 | At1g51820 | Probable LRR receptor-like serine/threonine-protein kinase At1g51820 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGW2 | PAM74 | Probable LRR receptor-like serine/threonine-protein kinase PAM74 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FN94 | At5g59670 | Receptor-like protein kinase At5g59670 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LIG2 | At3g21340 | Receptor-like protein kinase At3g21340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SNA3 | At3g46340 | Putative receptor-like protein kinase At3g46340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZQR3 | At2g14510 | Leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14510 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LPZ9 | SD113 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P43298 | TMK1 | Receptor protein kinase TMK1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGI2 | At1g67720 | Probable LRR receptor-like serine/threonine-protein kinase At1g67720 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2S4 | LECRKS4 | L-type lectin-domain containing receptor kinase S.4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWZ5 | SD25 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64556 | At2g19230 | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g19230 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SXB4 | At1g11300 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q84JQ4 | PRK2 | Pollen receptor-like kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q93ZS4 | NIK3 | Protein NSP-INTERACTING KINASE 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94AG2 | SERK1 | Somatic embryogenesis receptor kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64477 | At2g19130 | G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9XIC7 | SERK2 | Somatic embryogenesis receptor kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MERSLELLLL | LIRTLAIIHI | SQAQSQQGFI | SLDCGLPANE | PSPYTEPRTG | LQFSSDAAFI |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QSGKIGRIQA | NLEADFLKPS | TTMRYFPDGK | RNCYNLNVEK | GRNHLIRARF | VYGNYDGRDT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GPKFDLYLGP | NPWATIDLAK | QVNGTRPEIM | HIPTSNKLQV | CLVKTGETTP | LISVLEVRPM |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GSGTYLTKSG | SLKLYYREYF | SKSDSSLRYP | DDIYDRQWTS | FFDTEWTQIN | TTSDVGNSND |
| 250 | 260 | 270 | 280 | 290 | 300 |
| YKPPKVALTT | AAIPTNASAP | LTNEWSSVNP | DEQYYVYAHF | SEIQELQANE | TREFNMLLNG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KLFFGPVVPP | KLAISTILSV | SPNTCEGGEC | NLQLIRTNRS | TLPPLLNAYE | VYKVIQFPQL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ETNETDVSAV | KNIQATYELS | RINWQSDPCV | PQQFMWDGLN | CSITDITTPP | RITTLNLSSS |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GLTGTITAAI | QNLTTLEKLD | LSNNNLTGEV | PEFLSNMKSL | LVINLSGNDL | NGTIPQSLQR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KGLELLYQGN | PRLISPGSTE | TKSGKSFPVT | IVASVGSAAI | LIVVLVLVLF | LRKKKPSAVE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| VVLPRPSRPT | MNVPYANSPE | PSIEMKKRKF | TYSEVTKMTN | NFGRVVGEGG | FGVVCHGTVN |
| 610 | 620 | 630 | 640 | 650 | 660 |
| GSEQVAVKLL | SQSSTQGYKE | FKAEVDLLLR | VHHTNLVSLV | GYCDEGDHLA | LIYEFVPNGD |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LRQHLSGKGG | KPIVNWGTRL | RIAAEAALGL | EYLHIGCTPP | MVHRDVKTTN | ILLDEHYKAK |
| 730 | 740 | 750 | 760 | 770 | 780 |
| LADFGLSRSF | PVGGESHVST | VIAGTPGYLD | PEYYHTSRLS | EKSDVYSFGI | VLLEMITNQA |
| 790 | 800 | 810 | 820 | 830 | 840 |
| VIDRNRRKSH | ITQWVGSELN | GGDIAKIMDL | KLNGDYDSRS | AWRALELAMS | CADPTSARRP |
| 850 | 860 | 870 | 880 | ||
| TMSHVVIELK | ECLVSENSRR | NMSRGMDTLS | SPEVSMIFDA | EMIPRAR |