Q9FLW0
Gene name |
At5g24010 (MZF18.11) |
Protein name |
Probable receptor-like protein kinase At5g24010 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G24010 |
EC number |
|
Protein Class |
|
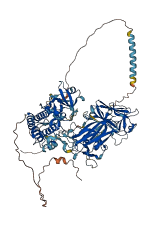
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
630-637 (Activation loop from InterPro)
Target domain |
489-764 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
648-655 (Activation loop from InterPro)
Target domain |
489-764 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9FLW0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9FLW0-F1 | Predicted | AlphaFoldDB |
52 variants for Q9FLW0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_5_8113919_C_A | 4 | P>T | No | 1000Genomes | |
| tmp_5_8113956_G_T | 16 | C>F | No | 1000Genomes | |
| tmp_5_8113970_C_T | 21 | L>F | No | 1000Genomes | |
| ENSVATH10971082 | 23 | F>L | No | 1000Genomes | |
| tmp_5_8114000_A_G | 31 | N>D | No | 1000Genomes | |
| ENSVATH10971083 | 32 | Y>H | No | 1000Genomes | |
| tmp_5_8114028_C_T | 40 | T>I | No | 1000Genomes | |
| ENSVATH10971135 | 41 | N>S | No | 1000Genomes | |
| ENSVATH00642399 | 49 | S>F | No | 1000Genomes | |
| ENSVATH03104723 | 55 | S>T | No | 1000Genomes | |
| ENSVATH03104724 | 56 | E>K | No | 1000Genomes | |
| tmp_5_8114081_G_A | 58 | G>S | No | 1000Genomes | |
| ENSVATH03104726 | 68 | I>N | No | 1000Genomes | |
| ENSVATH10971137 | 73 | T>P | No | 1000Genomes | |
| ENSVATH07025357 | 84 | N>K | No | 1000Genomes | |
| ENSVATH07025358 | 103 | G>D | No | 1000Genomes | |
| ENSVATH07025359 | 114 | F>I | No | 1000Genomes | |
| tmp_5_8114364_A_C | 152 | K>T | No | 1000Genomes | |
| tmp_5_8114378_G_A | 157 | V>I | No | 1000Genomes | |
| ENSVATH10971142 | 162 | F>V | No | 1000Genomes | |
| ENSVATH07025363 | 213 | V>I | No | 1000Genomes | |
| ENSVATH10971144 | 240 | Y>H | No | 1000Genomes | |
| tmp_5_8114640_G_A | 244 | R>K | No | 1000Genomes | |
| tmp_5_8114717_G_A | 270 | D>N | No | 1000Genomes | |
| tmp_5_8114899_G_T | 330 | E>D | No | 1000Genomes | |
| tmp_5_8114964_A_T | 352 | Y>F | No | 1000Genomes | |
| ENSVATH07025365 | 378 | A>E | No | 1000Genomes | |
| tmp_5_8115143_G_A | 412 | V>I | No | 1000Genomes | |
| ENSVATH07025367 | 440 | T>A | No | 1000Genomes | |
| ENSVATH00642401 | 441 | R>K | No | 1000Genomes | |
| tmp_5_8115239_G_A | 444 | E>K | No | 1000Genomes | |
| tmp_5_8115249_G_C | 447 | G>A | No | 1000Genomes | |
| tmp_5_8115286_T_G | 459 | N>K | No | 1000Genomes | |
| tmp_5_8115284_A_T | 459 | N>Y | No | 1000Genomes | |
| ENSVATH10971177 | 465 | R>G | No | 1000Genomes | |
| ENSVATH00642402 | 469 | S>T | No | 1000Genomes | |
| tmp_5_8115531_G_A | 541 | R>H | No | 1000Genomes | |
| tmp_5_8115537_G_A | 543 | R>H | No | 1000Genomes | |
| tmp_5_8115561_A_T | 551 | Y>F | No | 1000Genomes | |
| ENSVATH14151249 | 565 | M>I | No | 1000Genomes | |
| tmp_5_8115780_A_G | 624 | N>S | No | 1000Genomes | |
| tmp_5_8115889_A_T | 660 | E>D | No | 1000Genomes | |
| tmp_5_8116091_C_G | 728 | P>A | No | 1000Genomes | |
| ENSVATH07025376 | 728 | P>L | No | 1000Genomes | |
| tmp_5_8116098_C_T | 730 | S>L | No | 1000Genomes | |
| tmp_5_8116112_G_A | 735 | A>T | No | 1000Genomes | |
| ENSVATH14151251 | 745 | Y>C | No | 1000Genomes | |
| tmp_5_8116250_G_A | 781 | V>I | No | 1000Genomes | |
| tmp_5_8116260_C_T | 784 | P>L | No | 1000Genomes | |
| ENSVATH00642403 | 798 | E>G | No | 1000Genomes | |
| tmp_5_8116323_C_T | 805 | T>I | No | 1000Genomes | |
| ENSVATH10971180 | 808 | I>T | No | 1000Genomes |
No associated diseases with Q9FLW0
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| plasmodesma | A fine cytoplasmic channel, found in all higher plants, that connects the cytoplasm of one cell to that of an adjacent cell. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
40 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q07407 | htl | Fibroblast growth factor receptor homolog 1 | Drosophila melanogaster (Fruit fly) | PR |
| Q9Y616 | IRAK3 | Interleukin-1 receptor-associated kinase 3 | Homo sapiens (Human) | PR |
| P53667 | LIMK1 | LIM domain kinase 1 | Homo sapiens (Human) | PR |
| P53671 | LIMK2 | LIM domain kinase 2 | Homo sapiens (Human) | PR |
| Q06418 | TYRO3 | Tyrosine-protein kinase receptor TYRO3 | Homo sapiens (Human) | SS |
| P22607 | FGFR3 | Fibroblast growth factor receptor 3 | Homo sapiens (Human) | EV |
| P21802 | FGFR2 | Fibroblast growth factor receptor 2 | Homo sapiens (Human) | EV |
| P11362 | FGFR1 | Fibroblast growth factor receptor 1 | Homo sapiens (Human) | EV |
| P22455 | FGFR4 | Fibroblast growth factor receptor 4 | Homo sapiens (Human) | SS |
| P09619 | PDGFRB | Platelet-derived growth factor receptor beta | Homo sapiens (Human) | EV |
| P10721 | KIT | Mast/stem cell growth factor receptor Kit | Homo sapiens (Human) | EV |
| P16234 | PDGFRA | Platelet-derived growth factor receptor alpha | Homo sapiens (Human) | EV |
| P07333 | CSF1R | Macrophage colony-stimulating factor 1 receptor | Homo sapiens (Human) | SS |
| Q8K4B2 | Irak3 | Interleukin-1 receptor-associated kinase 3 | Mus musculus (Mouse) | PR |
| Q8CFA1 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Mus musculus (Mouse) | PR |
| O54785 | Limk2 | LIM domain kinase 2 | Mus musculus (Mouse) | PR |
| Q4QQS0 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Rattus norvegicus (Rat) | PR |
| O08680 | Epha3 | Ephrin type-A receptor 3 | Rattus norvegicus (Rat) | SS |
| P54757 | Epha5 | Ephrin type-A receptor 5 | Rattus norvegicus (Rat) | SS |
| P54759 | Epha7 | Ephrin type-A receptor 7 | Rattus norvegicus (Rat) | SS |
| P09759 | Ephb1 | Ephrin type-B receptor 1 | Rattus norvegicus (Rat) | SS |
| P0C0K7 | Ephb6 | Ephrin type-B receptor 6 | Rattus norvegicus (Rat) | SS |
| Q19238 | F09A5.2 | Putative tyrosine-protein kinase F09A5.2 | Caenorhabditis elegans | SS |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STF0 | LECRKS3 | Receptor like protein kinase S.3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LYN6 | At3g56050 | Probable inactive receptor-like protein kinase At3g56050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VZJ9 | CRCK2 | Calmodulin-binding receptor-like cytoplasmic kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94C25 | At5g20050 | Probable receptor-like protein kinase At5g20050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LTC0 | PBL19 | Probable serine/threonine-protein kinase PBL19 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FIL7 | CRCK1 | Calmodulin-binding receptor-like cytoplasmic kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ASQ5 | CRCK3 | Calmodulin-binding receptor-like cytoplasmic kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCZ4 | FER | Receptor-like protein kinase FERONIA | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAFPINLTQT | LLFFFCPLLH | LSFAAFTPTD | NYLINSGSNT | NTSFFTTRSF | LSDSSEPGSS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FLSTDRSISI | SDTNPSPDSP | VLYNTARVFP | VGGSYKFQVT | TKGTHFIRLH | FAPFKASRFN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LRSAKFRVLI | NGFSVINSFS | TSSVVVKEFI | LKIDDPVLEI | SFLPFKASGF | GFVNAVEVFS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| APKDYIMDQG | TKLVIPNSAQ | IFSNLSSQVL | ETVHRINVGG | SKLTPFNDTL | WRTWVVDDNY |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LLLRAAARRA | WTTHSPNYQN | GGATREIAPD | NVYMTAQEMD | RDNQELQARF | NISWGFQVDE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KRVLHLVRLH | FCDIVSSSLN | QLYFNVFINE | YLAFKDVDLS | TLTFHVLASP | LYIDFVAESD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RSGMLRISVG | PSDLSNPARV | NALLNGVEIM | RILSPVSSEV | VSGKRNVVWI | VVGSVLGGFV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| FLSLFFLSVL | CLCRRKNNKT | RSSESTGWTP | LRRFRGSSNS | RTTERTVSSS | GYHTLRISFA |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ELQSGTNNFD | RSLVIGVGGF | GMVFRGSLKD | NTKVAVKRGS | PGSRQGLPEF | LSEITILSKI |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RHRHLVSLVG | YCEEQSEMIL | VYEYMDKGPL | KSHLYGSTNP | PLSWKQRLEV | CIGAARGLHY |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LHTGSSQGII | HRDIKSTNIL | LDNNYVAKVA | DFGLSRSGPC | IDETHVSTGV | KGSFGYLDPE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| YFRRQQLTDK | SDVYSFGVVL | FEVLCARPAV | DPLLVREQVN | LAEWAIEWQR | KGMLDQIVDP |
| 730 | 740 | 750 | 760 | 770 | 780 |
| NIADEIKPCS | LKKFAETAEK | CCADYGVDRP | TIGDVLWNLE | HVLQLQESGP | LNIPEEDYGD |
| 790 | 800 | 810 | 820 | ||
| VTDPRTARQG | LSNGSNIERD | YGDGTSGIIS | STQVFSQLMT | NAGR |